- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Automated Tracking of Root for Confocal Time-lapse Imaging of Cellular Processes
(*contributed equally to this work) Published: Vol 7, Iss 8, Apr 20, 2017 DOI: 10.21769/BioProtoc.2245 Views: 11795
Reviewed by: Tie LiuShahin S. AliIsabelle Colas

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Near-Infrared Autofluorescence Imaging of Nuclei in Living Plant Roots
Akira Yoshinari and Masayoshi Nakamura
Apr 20, 2025 2132 Views

Direct Plant Regeneration From Immature Male Inflorescence of Banana (Musa spp.)
Pradeep Chand Deo
Oct 20, 2025 1437 Views

Live-Cell Monitoring of Piecemeal Chloroplast Autophagy
Masanori Izumi [...] Shinya Hagihara
Nov 5, 2025 1690 Views
Abstract
Here we describe a protocol that enables to automatically perform time-lapse imaging of growing root tips for several hours. Plants roots expressing fluorescent proteins or stained with dyes are imaged while they grow using automatic movement of the microscope stage that compensates for root growth and allows to follow a given region of the root over time. The protocol makes possible the image acquisition of multiple growing root tips, therefore increasing the number of recorded mitotic events in a given experiment. The protocol also allows the visualization of more than one fluorescent protein or dye simultaneously, using multiple channel acquisition. We particularly focus on imaging of cytokinesis in Arabidopsis root tip meristem, but this protocol is also suitable to follow root hair growth, pollen tube growth, and other regions of root over time, in various plant species. It may as well be amenable to automatically track non-plant structures with an apical growth.
Keywords: Cell divisionBackground
Cytokinesis is the last step of cell division, when the mother cell cytoplasm is partitioned between two daughter cells (Lipka et al., 2015). In plants, it is achieved through the centrifugal expansion of a cell plate in the division plane, which eventually becomes the newly synthetized cell wall between the cells that underwent mitosis (Buschmann and Zachgo, 2016; Müller and Jürgens, 2016). Plant cells, being embedded in a stiff cell wall, cannot migrate. Orientation of cell division together with elongation is therefore critical for organ morphogenesis. Root meristems are a good model to study cell division because they are easily amenable to microscopy techniques without the need of dissection. However, roots undergoing cell division grow in length, and therefore require manual adjustment of the observation field over time. This protocol allows easy time-lapse imaging of cytokinesis, and of other cellular processes.
Materials and Reagents
- 12-well microplates (Corning, Costar®, catalog number: 3513 )
- Microscope slides 76 x 26 x 1.1 mm (RS Components, catalog number: ISO 8037 )
- Microscope coverslips 22 x 60 mm (Thermo Fisher Scientific, Menzel-Gläser, catalog number: 630-2102 )
- Observation chambers, Lab-Tek II Chambered Coverglass W/Cover #1.5 Borosilicate (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 155360 )
- Strait scalpel blades (e.g., Swann Morton, straights mounted BS EN 27740 blades)
- Arabidopsis thaliana, 4 to 7 days-old seedlings of wild-type genotype, or expressing membrane and/or cell plate-localized fluorescent fusion proteins (e.g., 2xp35S::MP:YFP in Col-0, where MP is a myristoylation and palmitoylation signal sequence [Martinière et al., 2012; Simon et al., 2016]). Seedling can be grown vertically in squared plates (90 x 90 x 15) or round plates to get intact growing roots
- Murashige and Skoog (MS) medium (Sigma-Aldrich, catalog number: M5519 )
- Agar (Sigma-Aldrich, catalog number: A7921-1KG )
- FM4-64 (N-(3-Triethylammoniumpropyl)-4-(6-(4-(Diethylamino) Phenyl) Hexatrienyl) Pyridinium Dibromide) (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: T-3166 )
- Suitable fluorescent dyes (alternative) or fluorescent proteins for registration
- Plasma membrane or cell wall dyes such as FM4-64 or propidium iodide (Sigma-Aldrich, catalog number: P4864 )
- Nuclei dyes such as Hoechst (Thermo Fisher Scientific, InvitrogenTM, catalog number: H21486 )
- Plasma membrane or cell wall dyes such as FM4-64 or propidium iodide (Sigma-Aldrich, catalog number: P4864 )
Equipment
- Inverted confocal microscope (e.g., Carl Zeiss, model: AxioObserver Z1 ), equipped with a spinning disk module (e.g., Yokogawa Electric, model: CSU-W1 [T3 model])
- Metamorph and ImageJ software on the computer connected to the microscope
- 20 °C plant growth chamber with 24 h daylight (e.g., SANYO Electric, model: MLR-351 )
- Small tweezers (e.g., straight fine tweezers)
- Sterile hood
Software
- ImageJ (https://imagej.nih.gov/ij/; Schneider et al., 2012; this protocol was tested with version 1.49p but should be compatible with newer versions) with the plugins Turboreg (P. Thévenaz, http://bigwww.epfl.ch/thevenaz/turboreg/) and Stackreg (P. Thévenaz, http://bigwww.epfl.ch/thevenaz/stackreg/) installed are required. Fiji (http://fiji.sc/; Schindelin et al., 2012) could be used instead of ImageJ because it bundles the required plugins. To add the plugins, simply download them and copy them in ImageJ/plugins folder.
- Download the three files (CL_ini-Pos-Ch-finished.JNL; CL_ini-Pos-Ch.JNL; CL-root-track-MM_63multiD.ijm) attached as a zip file (named Files_BioProtocol_Doumane, as well available at this url http://www.ens-lyon.fr/RDP/SiCE/METHODS.html) and extract them in the folder C:\MM\app\mmproc\journal\root-tracking\. The ImageJ macro file CL-root-track-MM_63multiD.ijm should be edited according to the calibration of the objective used. Open the file in ImageJ (use the menu Open>File...), search for the line with ‘objective’, and replace the number 0.2063 with the real size of the pixel in microns, on this line and the next one (when using our 63x objective, 1 pixel is 0.2063 microns wide; Figure 1; Note 5). Save the file. Install the macro by selecting it in Plugins > Macro > Install. CL-root-track-MM_63multiD should appear in the Plugins > Macros > lower panel (using a 63x objective).
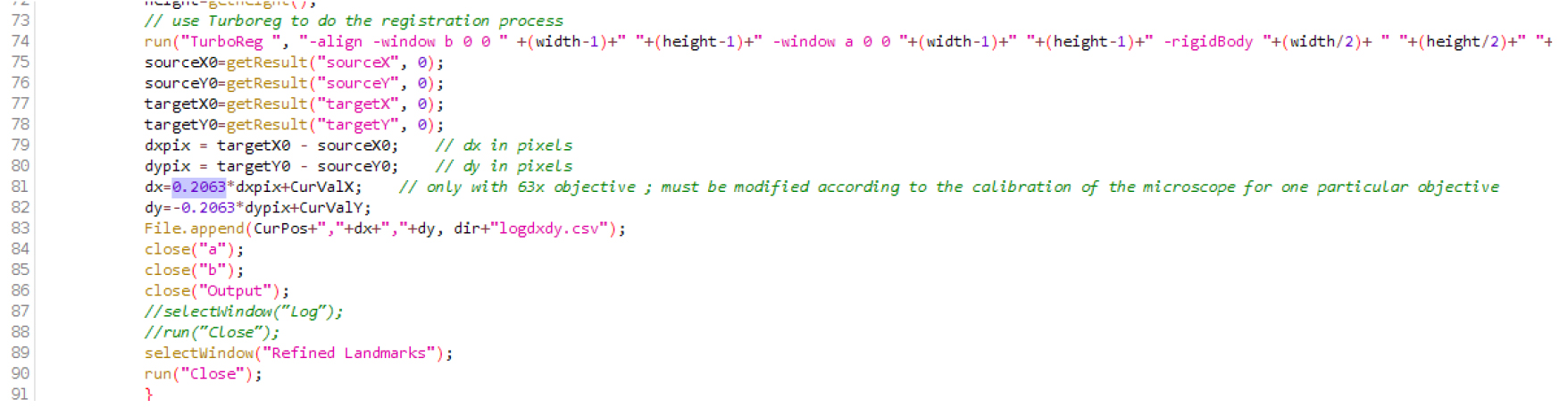
Figure 1. Macro file .ijm in the Fiji editor. Replace the number 0.2063 (here in lines 81 and 82) by the real size of the pixel (in microns) on your microscope, with the objective that will be used.
Procedure
- Automated imaging of growing Arabidopsis roots stained with a vital dye (see Video 1 and Figure 2)Video 1. Preparation of the Lab-Tek® observation chamber for FM4-64 staining. Pour 3 ml of ½ MS-0.8% agar with 5 μl of 100 mg/ml FM4-64 in the Lab-Tek® observation chamber.

Figure 2. Preparation of the Lab-Tek® observation chamber for FM4-64 staining. A. Drop 5 μl of 100 mg/ml FM4-64 into the observation chamber; B. Add 3 ml of pre-heated ½ MS-0.8% agar; C. Mix gently to homogenize; D. Let cool down and solidify.- Incubate seedlings in the FM4-64 bath (1 μg/ml FM4-64 in liquid ½ MS) for 5 to 10 min (time required for the preparation of the Lab-Tek® observation chamber).
- Prepare ½ MS-0.8% agar-FM4-64 in the Lab-Tek® observation chamber (see Recipes section and Video 2 and Figure 3). While seedlings incubate in the FM4-64 bath, pour ½ MS-0.8% agar-FM4-64 in the Lab-Tek® chamber, wait until it has solidified and dried.Video 2. Transfer of seedlings in the observation chamber. Cut out a piece of ½ MS-agar, add ½ MS and align carefully seedlings prior to putting back the squared piece of ½ MS-agar. Proceed to observation (FM4-64 staining) or let it stand several hours in a growth chamber so that seedlings acclimate to the Lab-Tek® chamber.
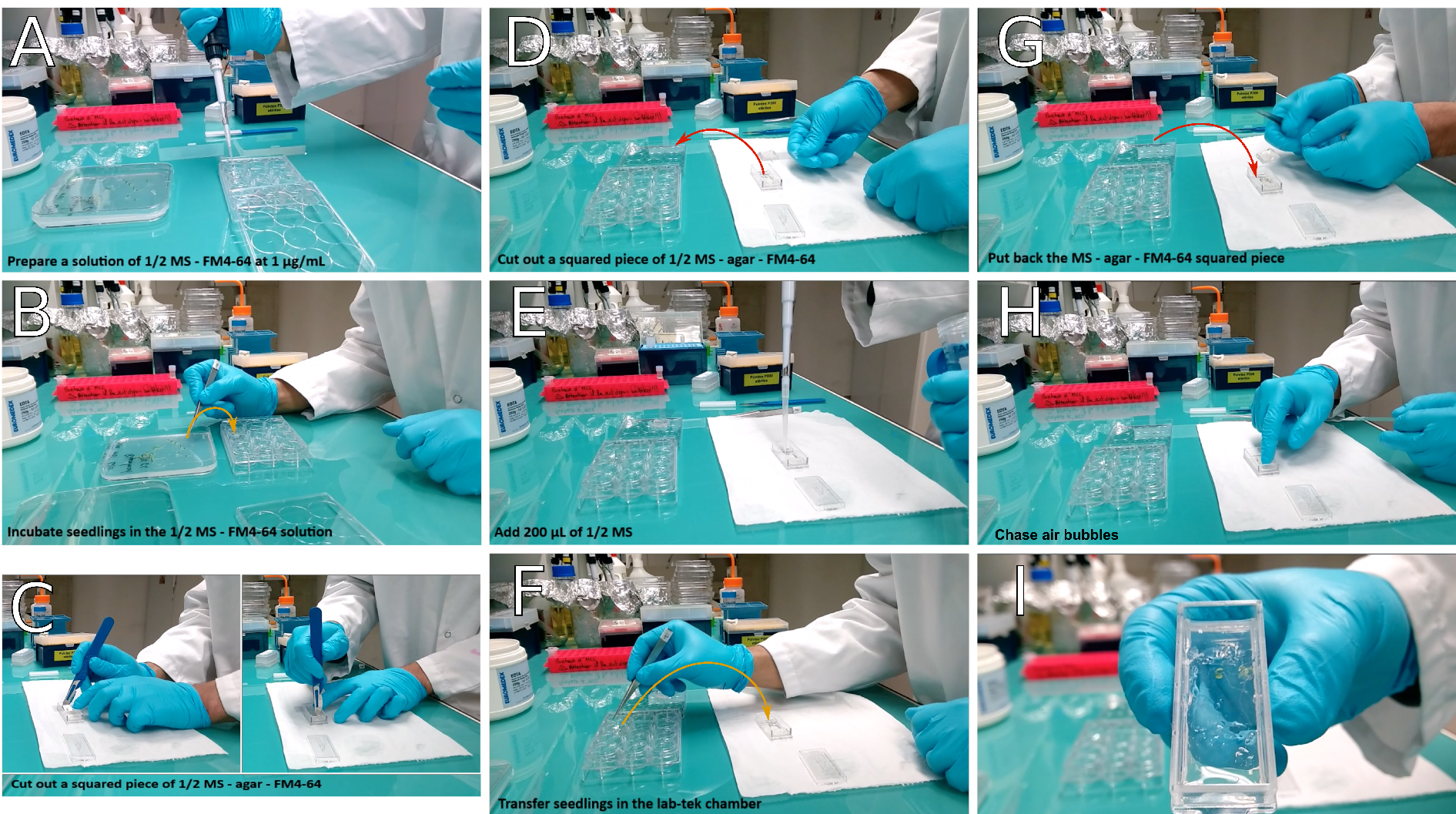
Figure 3. Transfer of seedlings in the observation chamber. If working without FM4-64, start the procedure at panel (C). In this case, prefer working under sterile conditions. A. Prepare a solution of ½ MS-FM4-64 at 1 μg/ml final concentration; B. Transfer the seedlings into the solution and incubate them for 5 min; C. In the meantime, cut a squared block of agar in the Lab-Tek® observation chamber; D. Then remove it from the chamber using a coverslips; E. Add 200 μl of ½ MS in the hole left in the chamber; F. Transfer the seedlings into the Lab-Tek® observation chamber. If you plan to leave Lab-Tek® observation chamber in a growth chamber overnight (when working without FM4-64), make sure seedlings are roughly aligned. G. Put the block of agar back inside the Lab-Tek® chamber, on top of either the whole seedlings, or only of their root system. H. Gently chase air bubbles with a gloved fingertip; I. Seedlings are ready for observation or to be grown overnight. - Cut a squared area of agar in the chamber. The piece of agar should go from one side of the chamber to the other, and be as wide as a coverslip (20 x 24 mm). Using a coverslip, remove the agar block from the observation chamber and keep it aside for later use (see step A6).
- Pour 200 µl of liquid ½ MS in the hole left by the removal of the agar block.
- Carefully align up to 4 seedlings in the Lab-Tek® observation chamber.
- Put back the cube of ½ MS-agar-FM4-64 on top of the seedling(s), chasing air bubbles manually (gently using a gloved fingertip).
- Place the sample under a microscope. Locate the first root tip by direct bright field observation under the microscope.
- Go to step B8.
- Incubate seedlings in the FM4-64 bath (1 μg/ml FM4-64 in liquid ½ MS) for 5 to 10 min (time required for the preparation of the Lab-Tek® observation chamber).
- Automated imaging of growing Arabidopsis roots expressing fluorescent proteins (see Naramoto et al., 2015; Aki and Umeda, 2016 for detailed alternatives procedure)
- Under the sterile hood, pour 3 ml of pre-heated ½ MS-0.8% agar in the observation chamber and let it cool down.
- Use the scalpel to cut a squared area of agar (20 x 24 mm) from one side to the other of the chamber, and as wide as a coverslip. Use a coverslip to remove the piece of agar from the observation chamber and keep it aside for later use (see step B5).
- Pour 200 µl of liquid ½ MS in the hole left by the removal of the agar block.
- Carefully align up to 4 seedlings in the observation chamber.
- Gently put back the cube of ½ MS-0.8% agar-FM4-64 on top of the seedling(s). Whole small/young seedlings can be positioned between the agar block and the bottom of the chamber. For longer/older seedlings, only the root system can be positioned between the agar and the bottom, with the aerial part of the seedling rising out of the agar. Chase air bubbles manually, gently using a gloved fingertip.
- Leave the plants to grow in their new environment, with an inclination of ± 70° of angle, in a growth chamber overnight, so that seedlings acclimate to the observation chamber and keep growing.
- Place the sample under a microscope. Locate the first root tip by bright field observation under the microscope.
- In Metamorph, use the menu ‘Apps’ > ‘Multi Dimensional Acquisition’ (MDA).
- In the ‘Main’ tab, choose the dimensions ‘Timelapse’, ‘Multiple Stage Positions’, ‘Multiple Wavelengths’ (even if you have only one position and/or one channel) and ‘Run journals’. These modalities are required. You can use the options ‘Z series’ and ‘Stream’ too if needed (Figure 4): these modalities are optional, but compatible with each other.
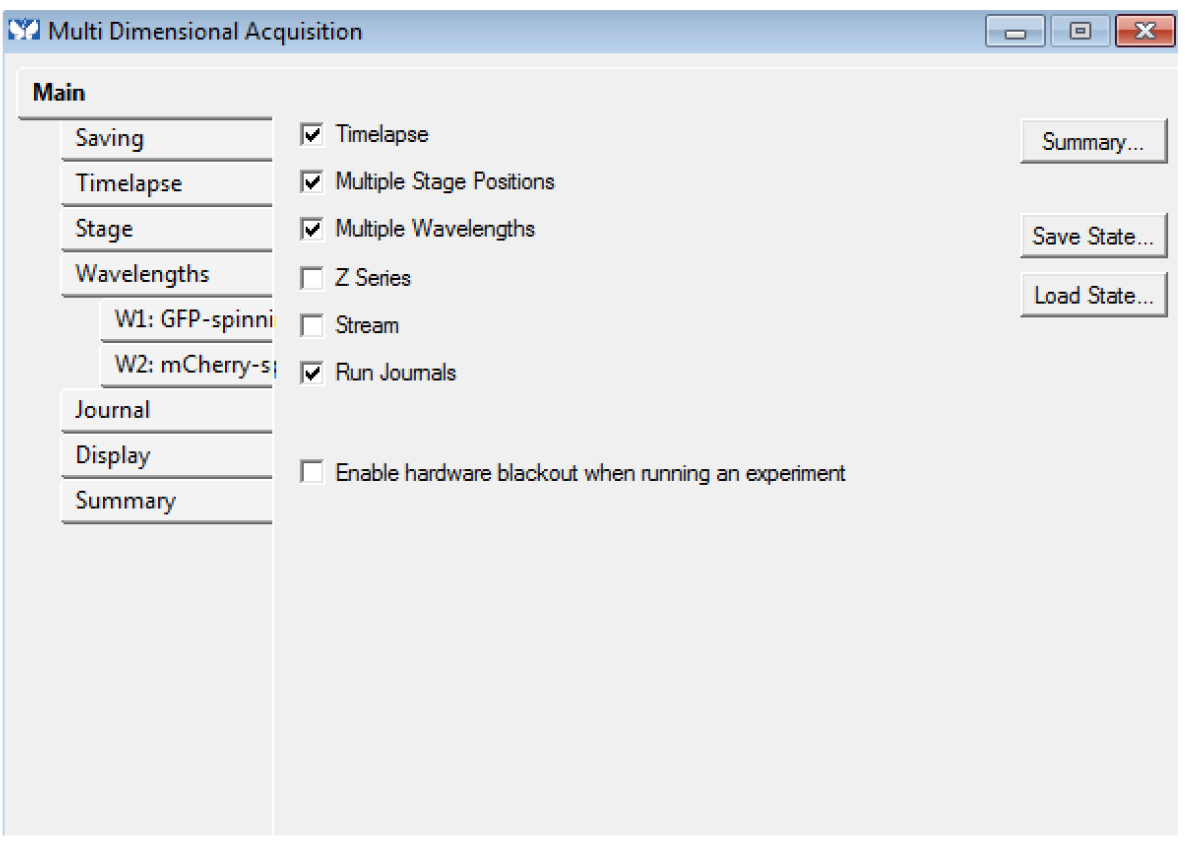
Figure 4. The Multi Dimensional Acquisition Main tab: Timelapse, Multiple Stage Positions (even for only one), Multiple Wavelengths (even for only one) and Run Journals are required. Z series and Stream are optional. - Choose an empty folder to save the images. You must not use ‘_w’ or ‘_s’ in the base name of the images.
- Change the other settings of the selected tabs of the MDA menu as needed (use the same settings as for a regular time-lapse acquisition): you could use the settings displayed in Figure 5 as a starting point. If you use the ‘Z series’ option, make sure you remember slice number that should be used for the registration (we use the central slice of the stack most of the time; Figure 5).
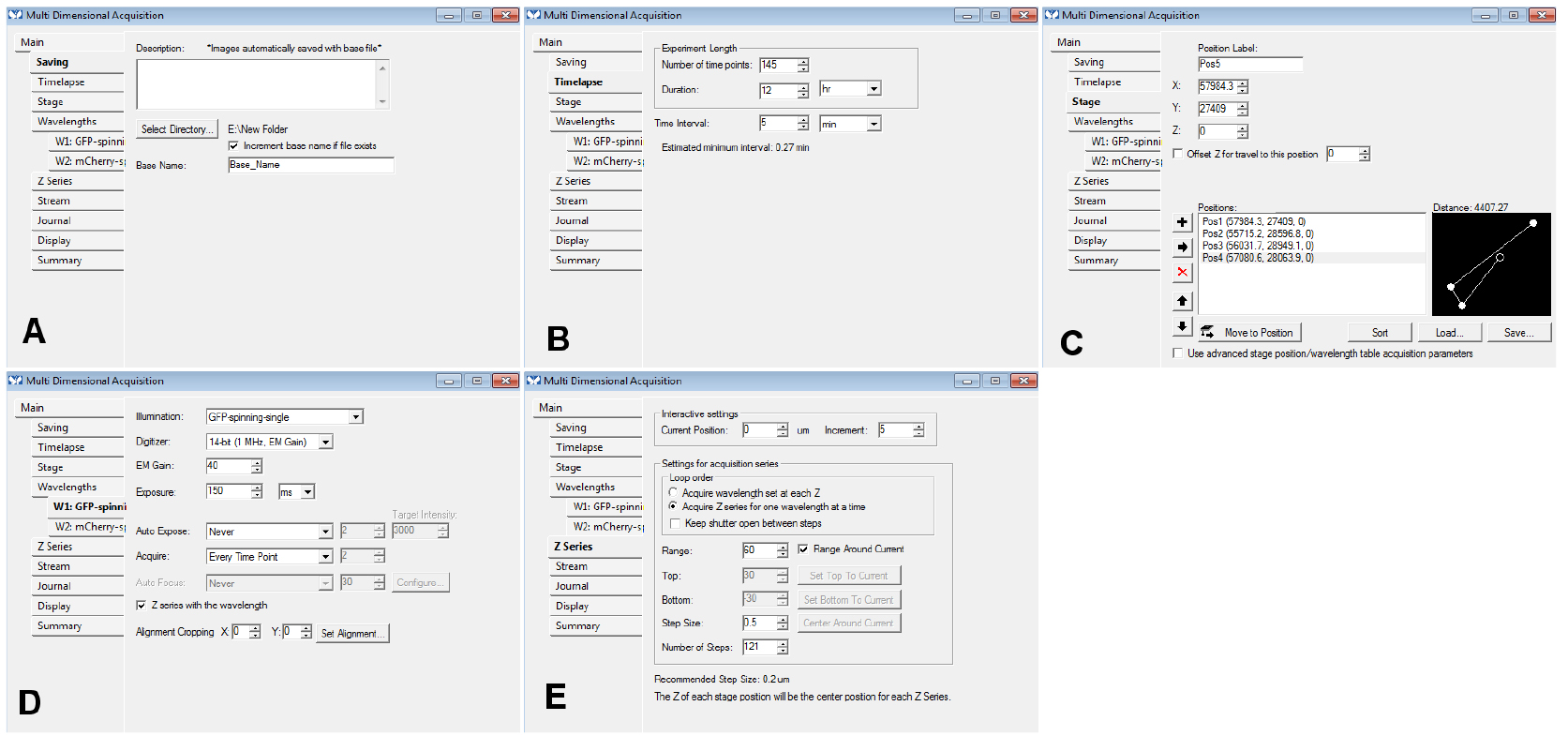
Figure 5. An example of the settings we often use for the tabs of the MDA module. In the Z series, make sure you remember the number of steps (here, 121) in E: if you want to use the central slice for the registration, you need to use the number 60 for the ‘Slice used for registration’ in step B15. - In the ‘Journal’ menu, choose ‘CL_ini-Pos-Ch.jnl’ for ‘end of stage position’, ‘CL_ini-Pos-Ch-finished.jnl’ for ‘end of acquisition’, ‘CL_ini-Pos-Ch-finished.jnl’ for ‘on cancel’ (Figure 6).
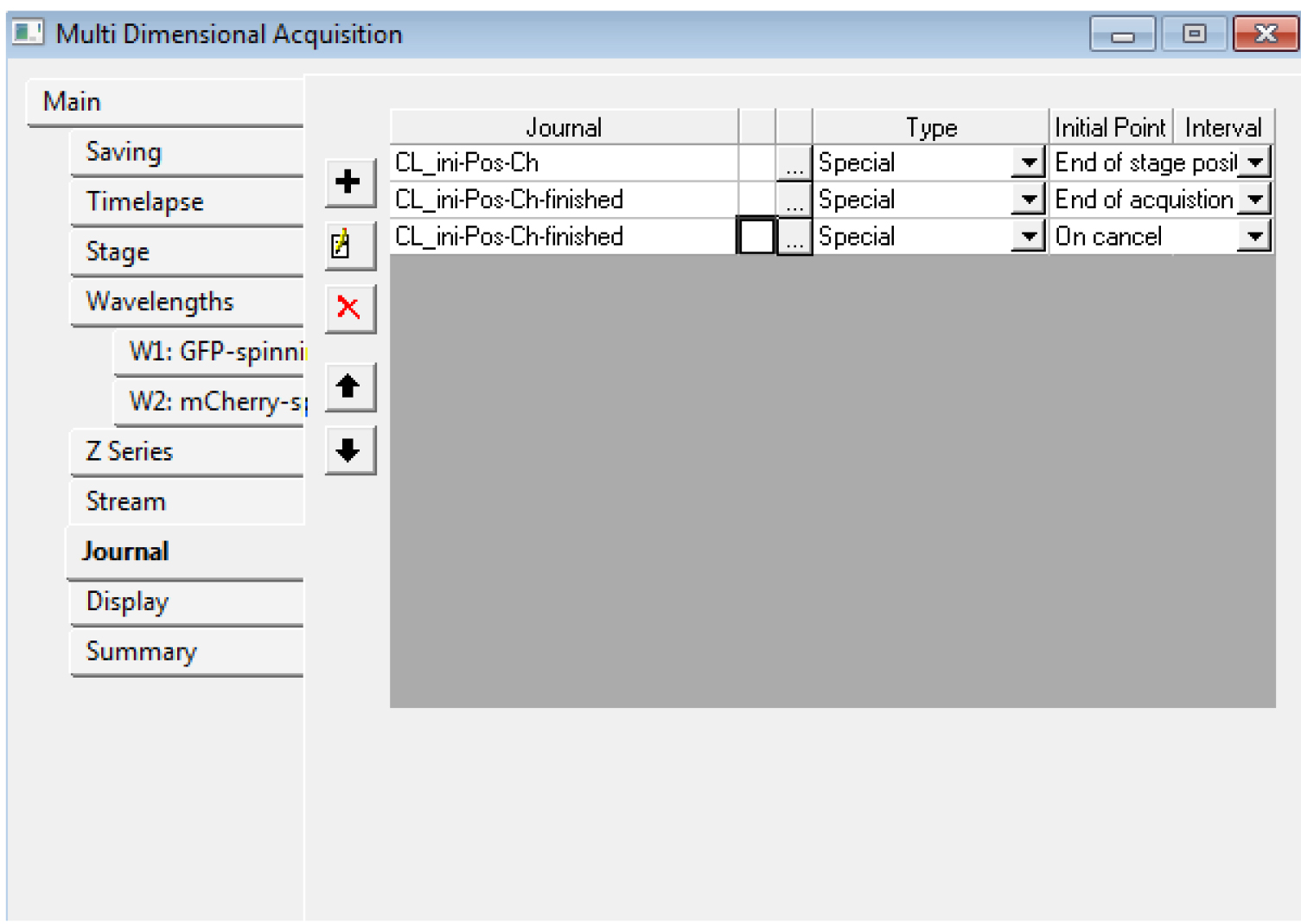
Figure 6. The Multi Dimensional Acquisition Journal tab. Choose the proper journal for each category. - You should save the settings of the MDA in a file (‘save state...’ in the Main tab) for the next time. Do not start the acquisition with Metamorph yet.
- Start ImageJ/Fiji and run the macro CL-root-track-MM_63multiD.ijm.
- A window pops up. Fill in the ‘Number of positions’ (as in the MDA, 1 or more), the ‘Channel used for registration’ (the channel number in the MDA, 1 or more), and the ‘Slice used for registration’ (as explained in step B11; Figure 7)
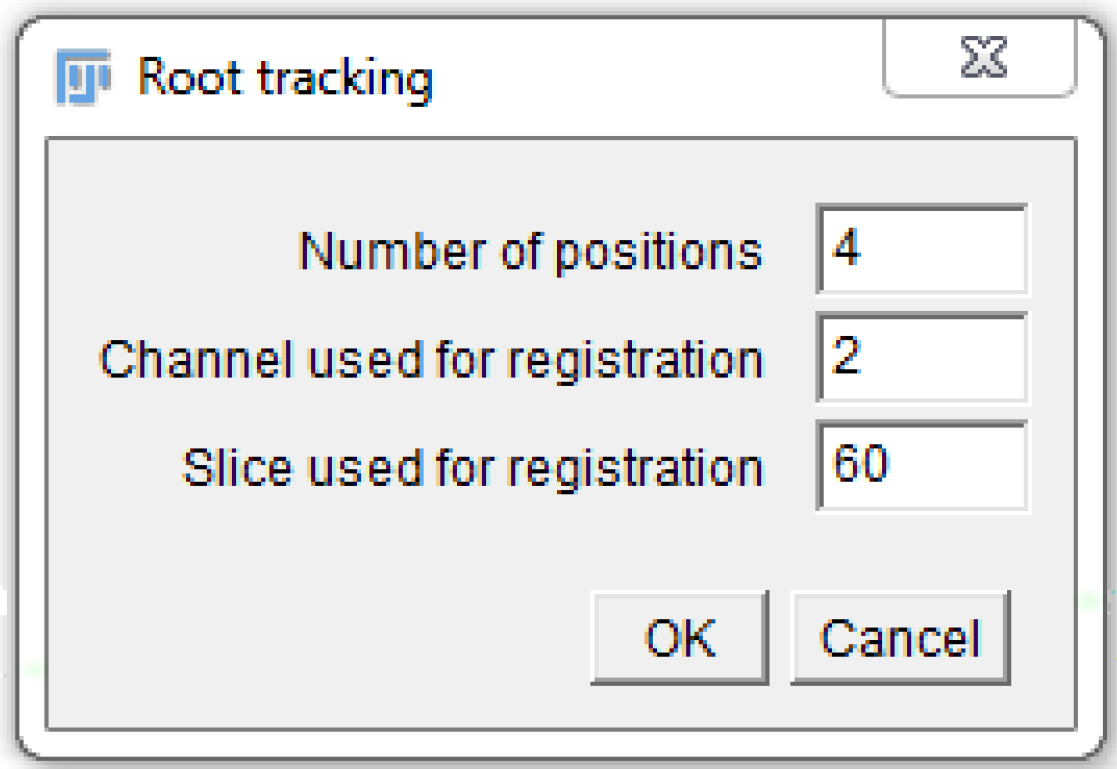
Figure 7. Fill in the macro pop-up window with the appropriate numbers, according to the settings chosen in the MDA module. Here is an example that is compatible with Figure 5. - Another window pops up. Select the folder used in the MDA to save the images.
- Do not close ImageJ or any of its opened windows. Go back to Metamorph. Select in the menu ‘Journal’ > ‘Journal control’ > ‘Stopwatches’: let the new window opened.
- Start the acquisition with the ‘Acquire’ button in the MDA (see Note 4).
- Under the sterile hood, pour 3 ml of pre-heated ½ MS-0.8% agar in the observation chamber and let it cool down.
Data analysis
After making a movie of your stacks, you obtain stable image over time (Video 3).
Notes
- You can check that X and Y coordinates change overtime for each root, to make sure registration is occurring. Variations in positions should be observed from the third time-point.
- The suitability of the dye for relative position registration could be tested with short regular time-lapse acquisition (e.g., 3-5 time-points and without real-time tracking): if the plugin Stackreg is able to correctly register the slices of this acquisition, we can consider that the dye is appropriate. It is also a good way to test the correct interval of time to use to do the time-lapse acquisition.
- If you want to perform long time-lapse analysis (e.g., 4-24 h), it is better to work in sterile conditions under a laminar flow hood. For short time lapses, this is optional.
- At the end of the acquisition, you will have the images or stacks, with the roots that will seem almost stationary over the time (a registration post-acquisition could be necessary, if the movement of the root was not linear enough). You will find a csv file with the displacements of the stage (in microns) recorded as a log, that you will need if you want to measure the speed of objects in the image, or if you need to troubleshoot the process. If a problem appears with ImageJ during the acquisition, Metamorph will continue to do the acquisition, and will use the last calculated displacements in X and Y for all the next time points (no additional coordinates will be recorded in the csv file).
- This protocol has been tested on our microscope with objective's magnifications ranging from 10x to 63x. But there is no limitation on this factor, as long as the macro file is edited as described in the second paragraph of the ‘Software’ section.
Recipes
- FM4-64 stock solution
- Resuspend FM4-64 powder in sterile distilled water and make a stock solution at 100 mg/ml (add 100 μl of sterile distilled water to 100 μg of FM4-64 powder)
- FM4-64 can be stored away from light at -20 °C as a powder, and up to 3 months at 100 mg/ml at -20 °C
- ½ MS-FM4-64 bath
- In 12 wells microplates, dilute FM4-64 stock solution to 1 μg/ml final concentration in 1 ml of half strength MS (½ MS)
- Mix gently
- Lab-Tek® chamber with ½ MS-0.8% agar-FM4-64 dye (see Video 2)
- Prepare ½ MS-0.8-% agar medium and let it cool down to ± 60 °C
- Under a sterile hood, drop 5 μl of FM4-64 stock solution in the observation chamber (see Note 3)
- Add 3 ml of ½ MS-0.8% agar medium and mix gently
- Let it cool down until it solidifies
- Prepare ½ MS-0.8-% agar medium and let it cool down to ± 60 °C
Acknowledgments
We thank Dr. Laia Armengot and Dr. Antoine Larrieu for critical comments on the manuscript. M.D. is funded by a fellowship from the French Ministry of Higher Education and Research, Y.J. by ERC No. 3363360-APPL under FP/2007-2013, M-C.C by a group leader starting package «fond de recherche» from ENS Lyon.
References
- Aki, S. S. and Umeda, M. (2016). Cytrap marker systems for in vivo visualization of cell cycle progression in Arabidopsis. Methods Mol Biol 1370: 51-57.
- Buschmann, H. and Zachgo, S. (2016). The evolution of cell division: from streptophyte algae to land plants. Trends Plant Sci 21(10): 872-883.
- Federici, F., Dupuy, L., Laplaze, L., Heisler, M. and Haseloff, J. (2012). Integrated genetic and computation methods for in planta cytometry. Nat Methods 9(5): 483-485.
- Larrieu, A., Champion, A., Legrand, J., Lavenus, J., Mast, D., Brunoud, G., Oh, J., Guyomarc’h, S., Pizot, M., Farmer, E. E., Turnbull, C., Vernoux, T., Bennett, M. J. and Laplaze, L. (2015). A fluorescent hormone biosensor reveals the dynamics of jasmonate signalling in plants. Nat Commun 6: 6043.
- Lipka, E., Herrmann, A. and Mueller, S. (2015). Mechanisms of plant cell division. Wiley Interdiscip Rev Dev Biol 4(4): 391-405.
- Martinière, A., Lavagi, I., Nageswaran, G., Rolfe, D. J., Maneta-Peyret, L., Luu, D. T., Botchway, S. W., Webb, S. E., Mongrand, S., Maurel, C., Martin-Fernandez, M. L., Kleine-Vehn, J., Friml, J., Moreau, P. and Runions, J. (2012). Cell wall constrains lateral diffusion of plant plasma-membrane proteins. Proc Natl Acad Sci U S A 109(31): 12805-12810.
- Müller, S. and Jürgens, G. (2016). Plant cytokinesis-No ring, no constriction but centrifugal construction of the partitioning membrane. Semin Cell Dev Biol 53: 10-18.
- Naramoto, S., Dainobu, T. and Otegui, M. (2015). A bioimaging pipeline to show membrane trafficking regulators localized to the golgi apparatus and other organelles in plant cells. Bio-protocol 5(17): e1583.
- Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B., Tinevez, J. Y., White, D. J., Hartenstein, V., Eliceiri, K., Tomancak, P. and Cardona, A. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7): 676-682.
- Schneider, C. A., Rasband, W. S. and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9(7): 671-675.
- Simon, M. L., Platre, M. P., Marques-Bueno, M. M., Armengot, L., Stanislas, T., Bayle, V., Caillaud, M. C. and Jaillais, Y. (2016). A PtdIns(4)P-driven electrostatic field controls cell membrane identity and signalling in plants. Nat Plants 2: 16089.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Doumane, M., Lionnet, C., Bayle, V., Jaillais, Y. and Caillaud, M. (2017). Automated Tracking of Root for Confocal Time-lapse Imaging of Cellular Processes. Bio-protocol 7(8): e2245. DOI: 10.21769/BioProtoc.2245.
Category
Plant Science > Plant developmental biology > Morphogenesis
Cell Biology > Cell imaging > Confocal microscopy
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












