- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Single Molecule RNA FISH in Arabidopsis Root Cells
Published: Vol 7, Iss 8, Apr 20, 2017 DOI: 10.21769/BioProtoc.2240 Views: 16149
Reviewed by: Gal HaimovichAnca Flavia SavulescuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Novel Method to Map Small RNAs with High Resolution
Kun Huang [...] Jeffrey L. Caplan
Aug 20, 2021 4681 Views

Quantitative Analysis of RNA Editing at Specific Sites in Plant Mitochondria or Chloroplasts Using DNA Sequencing
Yang Yang and Weixing Shan
Sep 20, 2021 3292 Views

Profiling of Single-cell-type-specific MicroRNAs in Arabidopsis Roots by Immunoprecipitation of Root Cell-layer-specific GFP-AGO1
Lusheng Fan [...] Xuemei Chen
Dec 20, 2022 2231 Views
Abstract
Methods that allow the study of gene expression regulation are continually advancing. Here, we present an in situ hybridization protocol capable of detecting individual mRNA molecules in plant root cells, thus permitting the accurate quantification and localization of mRNA within fixed samples (Duncan et al., 2016; Rosa et al., 2016). This single molecule RNA fluorescence in situ hybridization (smFISH) uses multiple single-labelled oligonucleotide probes to bind target RNAs and generate diffraction-limited signals that can be detected using a wide-field fluorescence microscope. We adapted a recent version of this method that uses 48 fluorescently labeled DNA oligonucleotides (20 mers) to hybridize to different portions of each transcript (Raj et al., 2008). This approach is simple to implement and has the advantage that it can be readily applied to any genetic background.
Keywords: Single RNA moleculesBackground
While single molecule FISH has been developed to quantitatively measure mRNAs at the single cell level for cultured cells, tissue sections and whole-mount invertebrate organisms, this method was not optimized for use in single cells in plants. Fluorescence imaging in plants is considerably challenging due to endogenous autofluorescence of plant tissues. Here, we report a method to detect single RNA molecules in plants. We describe the detection and automated counting of single transcripts within cells of fixed Arabidopsis root squashes. This method generates isolated cells and single-cell layers, which together with the use of red and far-red dyes maximizes signal-to-noise ratio limiting background noise.
Materials and Reagents
- 1.5 ml microcentrifuge tube
- Sterile SterilinTM 10 cm square Petri dishes for plant growth media (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 109 )
- 22 x 22 mm No. 1 glass coverslips (VWR, catalog number: 631-0124 )
- Poly-L-Lysine slides (Sigma-Aldrich, catalog number: P0425 )
- Razor blades (Agar Scientific, catalog number: AGT586 )
- Parafilm M® sealing film (Bemis, catalog number: PM992 )
- Hybridization chamber
Note: Although these are available commercially (Corning, catalog number: 2551 ) we used 10 cm square Petri dishes covered externally with a layer of black insulation tape (RS Components, catalog number: 494-405 ). A double layer of tissue (KCWW, Kimberly-Clark, catalog number: 7557 ) was then placed in the base and saturated with sterile water before slides were placed on top of a single layer of Parafilm (see Figure 1).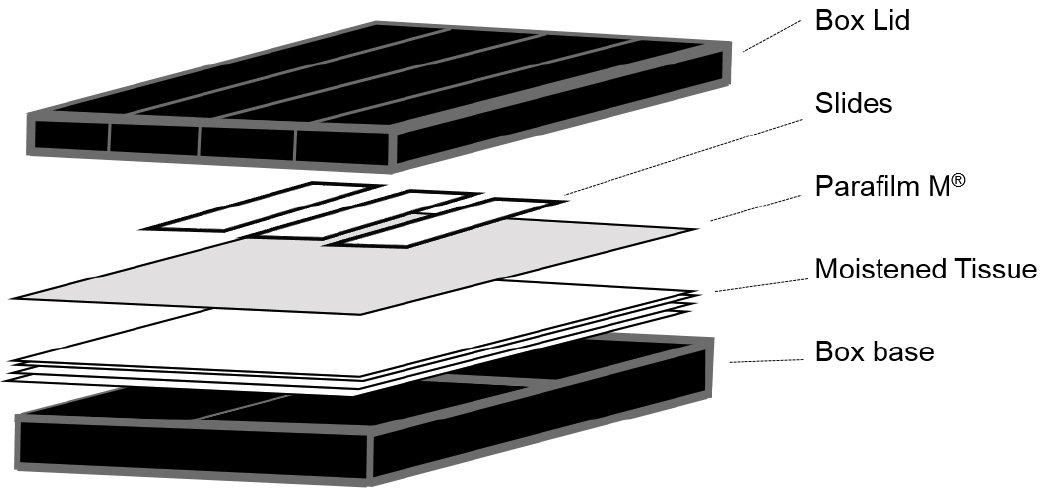
Figure 1. Illustration of the hybridization chambers used for smFISH experiments - Low stender-form preparation dishes (VWR, catalog number: 470144-866 ; or similar rimmed glass dish)
- Arabidopsis thaliana roots
- Sodium hypochlorite (NaClO) (VWR, BDH®, catalog number: CABDH7038-4L )
- Sucrose (Sigma-Aldrich, catalog number: S9378-1KG )
- Phytagel (Sigma-Aldrich, catalog number: P8169 )
- Paraformaldehyde (Sigma-Aldrich, catalog number: P6148 )
- Nuclease-free phosphate buffered saline solution (PBS, 10x) pH 7.4 (Thermo Fisher Scientific, AmbionTM, catalog number: AM9624 )
- Liquid nitrogen
- Ethanol suitable for molecular biology
- Nuclease-free 20x saline-sodium citrate (20x SSC) (Thermo Fisher Scientific, AmbionTM, catalog number: AM9763 )
- Clear nail varnish (Electron Microscopy Sciences, catalog number: 72180 ; or similar)
- Dextran sulphate (Sigma-Aldrich, catalog number: RES2029D )
- Custom RNA FISH Stellaris® probe sets
We used the online Stellaris Probe Designer to design our smFISH probe sets: https://www.biosearchtech.com/support/education/stellaris-rna-fish. We used a default masking level 2 to avoid general problematic RNA sequences and selected the maximum number of 48 probes, an oligo length of 20 nt and 2 nt minimum spacing between probes.
We also performed TAIR BLAST searches (https://www.arabidopsis.org/Blast/index.jsp) for each probe sequence and considered the results collectively to ensure target specificity. We found Quasar 570 and Quasar 670 dyes equally suitable for imaging RNA in Arabidopsis root cells, however we were unable to detect RNA labelled with Fluorescein modified probes. - Murashige & Skoog basal medium with vitamins (any equivalent source would be suitable) (PhytoTechnology Laboratories®, catalog number: M519 )
- Nuclease-free Tris-EDTA buffer solution (10 mM Tris-HCl, 1 mM EDTA pH 8) (Sigma-Aldrich, catalog number: 93283 )
- DAPI (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: D1306 )
- Glucose (Sigma-Aldrich, catalog number: 158968-500G )
- Tris HCl buffer 1 M pH 8, nuclease-free (Thermo Fisher Scientific, AmbionTM, catalog number: AM9855G )
- Glucose oxidase (Sigma-Aldrich, catalog number: G0543 )
- Bovine liver catalase (Sigma-Aldrich, catalog number: C3155 )
- Deionized formamide (Sigma-Aldrich, catalog number: F9037 )
- 1 N HCl (Sigma-Aldrich, catalog number: 71763 )
- 10% (v/v) bleach (Vortex 5-10% sodium hypochlorite, Procter & Gamble, UK) diluted in dH2O
- TE buffer (10 nM Tris-HCl, 1 mM EDTA, pH 8.0)
- Nuclease-free water - not DEPC treated (QIAGEN, catalog number: 129117 )
- Murashige and Skoog medium (see Recipes)
- Wash buffer (see Recipes)
- Hybridization solution (see Recipes)
- DAPI solution (see Recipes)
- Anti-fade GLOX buffer (minus enzymes) (see Recipes)
- Anti-fade GLOX buffer (containing enzymes) (see Recipes)
Equipment
- Forceps (Fisher Scientific, S MurrayTM, catalog number: E017/01 ; or similar)
- Coplin jar (Sigma-Aldrich, catalog number: S6016 ; or similar)
- Plant growth chamber (Panasonic Healthcare, Sanyo, catalog number: MLR-352H-PE ; or similar)
- Fume cupboard (Labconco, catalog number: 2247300 ; or similar)
- Laminar flow cabinet (Thermo Fisher Scientific, Thermo ScientificTM, model: HeraguardTM ECO Clean Bench Floor Stands , catalog number: 50109313; or similar)
- Laboratory oven (Cole-Palmer, catalog number: WZ-52412-83 ; or similar)
- Orbital shaker (Cole-Palmer, catalog number: WZ-51700-13 ; or similar)
- Wide-field fluorescence microscope
Note: We used an Andor iXon EM-CCD camera (Andor, model: iXon EMCCD Camera ) fitted to a Zeiss, Elyra PS-1 (Zeiss, model: Elyra PS.1 ), however similar images have also been obtained using a standard CCD camera optimized for low light level imaging. - A high numerical aperture (> 1.3) and 60 or 100x oil-immersion objective
- Strong light source, such as a mercury or metal-halide lamp (Xenon or LEDs are typically not bright enough)
- Filter sets appropriate for the fluorophores
Software
- ImageJ software: http://rsbweb.nih.gov/ij/index.html (Schindelin et al., 2015)
- FISHcount: https://github.com/JIC-CSB/FISHcount
- Graphpad Prism Software or MS Excel
Procedure
- Plant growth and microscope slide preparation
- Surface sterilize seeds in 5% (v/v) sodium hypochlorite for 5 min in a 1.5 ml microcentrifuge tube (inverting ~10 times) and then rinse three times in sterile distilled water.
- Plate the seeds on to Murashige and Skoog medium (pH 5.8) supplemented with 1% (w/v) sucrose and 0.5% (w/v) phytagel.
- Stratify the seeds by incubating for 2 days at 4 °C in darkness. Then, seedlings were grown at 22 ± 1 °C, in vertically oriented Petri dishes for at least 5 days (16 h light and 8 h dark).
- Surface sterilize seeds in 5% (v/v) sodium hypochlorite for 5 min in a 1.5 ml microcentrifuge tube (inverting ~10 times) and then rinse three times in sterile distilled water.
- Fixation and preparation of Arabidopsis roots (see Figure 2 for an overview)
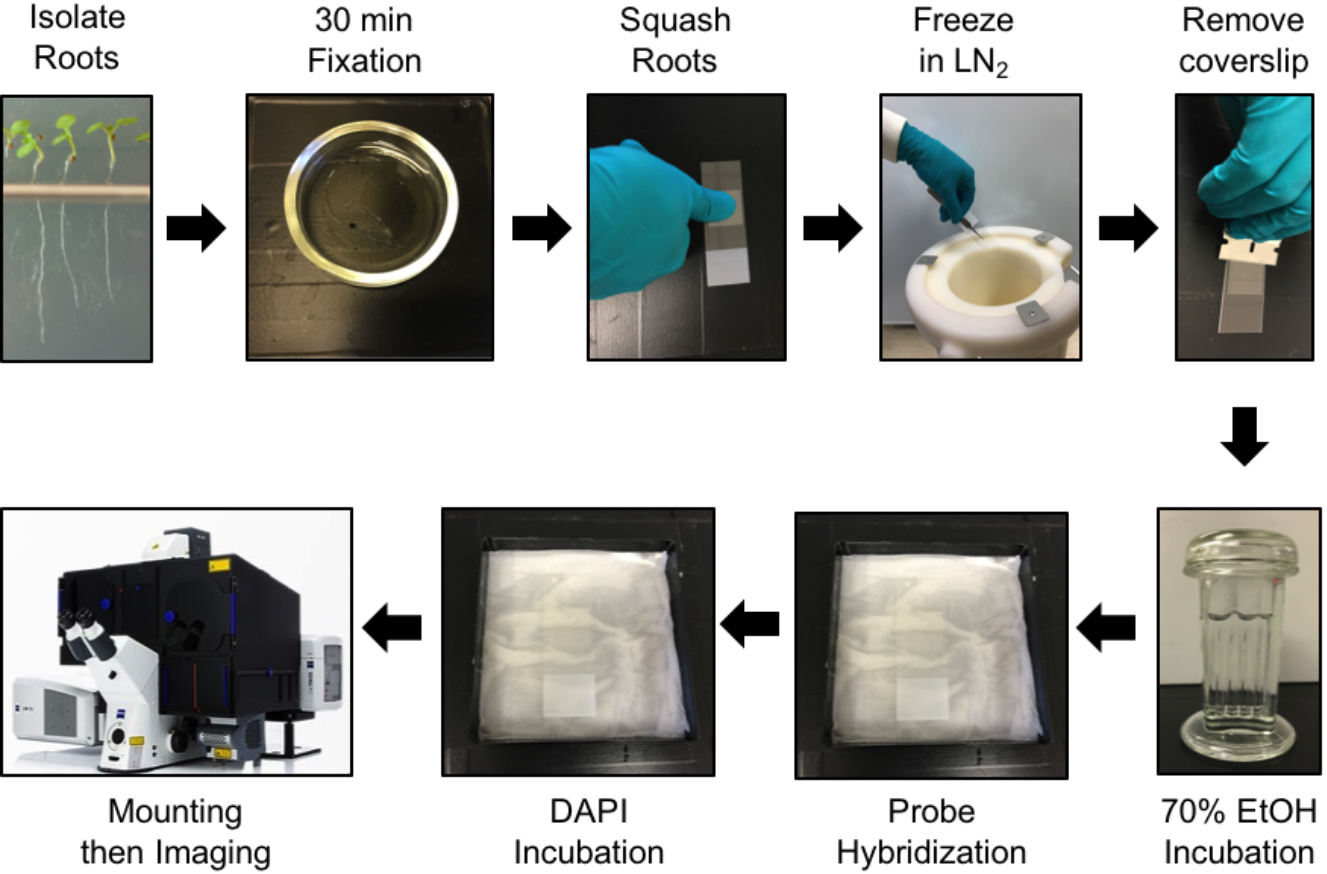
Figure 2. Protocol steps for smFISH in Arabidopsis root cells. See main text for details.
- Cut root tips from Arabidopsis seedlings (approx. 1 cm) whilst still on plates and place into a small glass dish containing freshly prepared 4% paraformaldehyde and incubate for 30 min at room temperature under a fume hood.
- Remove the roots from the fixative and wash twice with 1x PBS.
- Arrange 3-4 roots onto a slide and cover with a coverslip. Gently squash each root onto the slide using either your thumb and be careful to avoid breaking the coverslip. Aim to splay the roots sufficiently to produce multiple files and isolated cells in a single cell layer (see Figure 3).
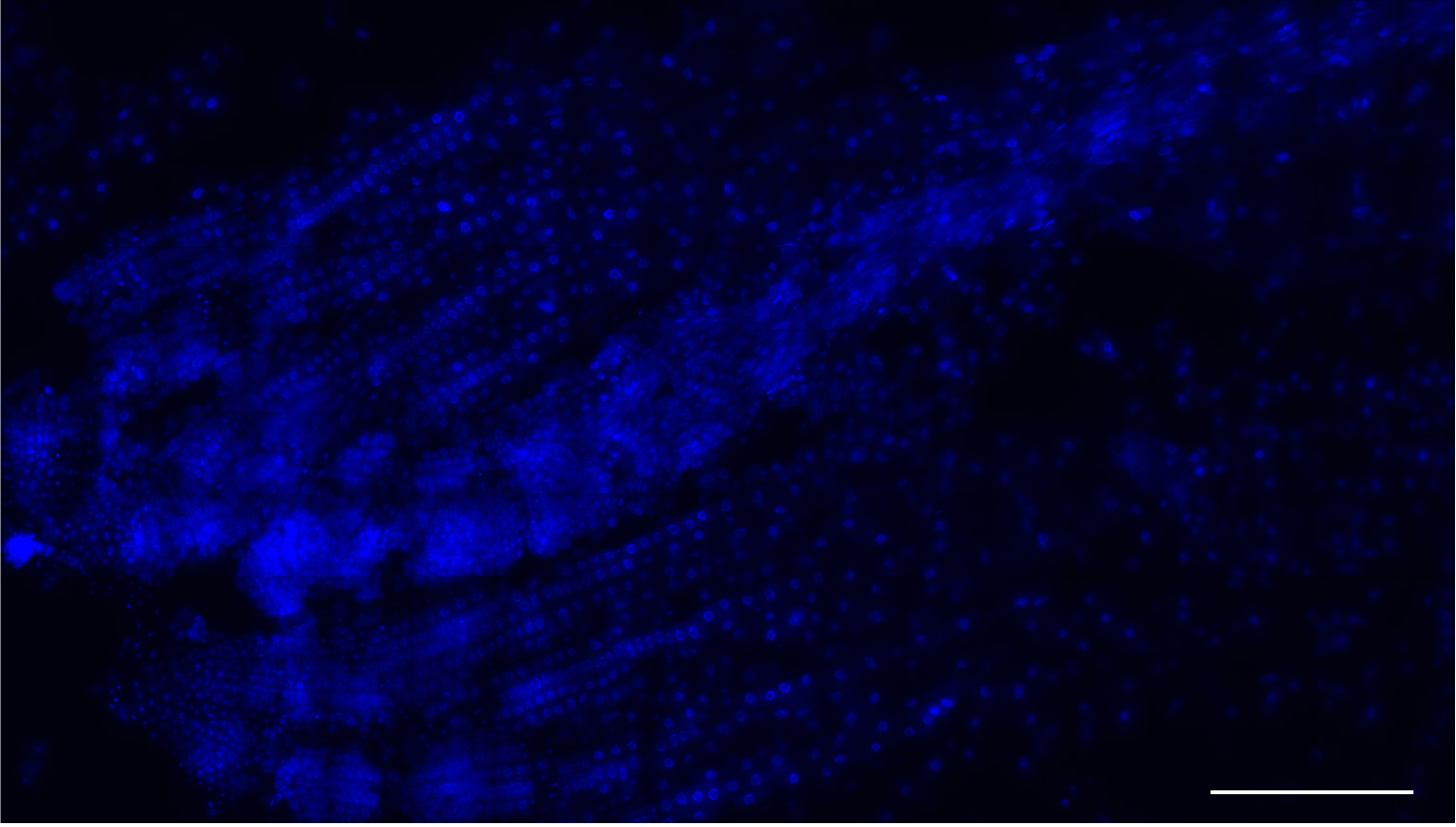
Figure 3. Low magnification image a root meristem squash. Nuclei stained with DAPI (blue). This method yields many single-layer cells that are either isolated or within files. Scale bar = 200 μm. - Use tweezers to hold the squashed roots under the coverslip and immerse each slide in liquid nitrogen for ~5 sec. After removal from the nitrogen, ease a razor blade between the coverslip and the slide and flick the coverslip off.
- Leave samples to air dry at room temperature for a minimum of 30 min.
Note: To avoid increased levels of autofluorescence do not leave to dry for longer than 2 h. - Permeabilize the samples by immersing the slides into a Coplin jar containing 70% ethanol for a minimum of one hour at room temperature. Fixed roots can be stored at 2 to 8 °C in 70% ethanol up to a week prior to hybridization.
- Cut root tips from Arabidopsis seedlings (approx. 1 cm) whilst still on plates and place into a small glass dish containing freshly prepared 4% paraformaldehyde and incubate for 30 min at room temperature under a fume hood.
- Probe hybridization
- Remove slides from the ethanol and allow residual ethanol to evaporate at room temperature.
- Wash 2 times on the slide with wash buffer for 5 min.
- Prepare a probe solution by adding 1 μl of each required probe stock solution to 100 μl of hybridization solution (250 nM final concentration).
- Add 100 μl probe solution to each slide, cover with a coverslip to prevent evaporation and incubate in a humid chamber at 37 °C for a minimum of 4 h (or overnight) in the dark.
- Remove slides from the ethanol and allow residual ethanol to evaporate at room temperature.
- Sample mounting
- Remove the cover slip and wash the samples 2 times in 200 μl wash buffer for 5 min on the slide.
- Immerse the slide in a Coplin jar containing ~30 ml of wash buffer and incubate for 30 min at 37 °C, in the dark.
- Remove the slide from the jar and add 100 μl of DAPI solution to the slide then incubate at 37 °C for 30 min in the dark.
- Aspirate the DAPI solution carefully with a pipette and rinse with ~100 μl 2x SSC.
- Remove 2x SSC and add 100 μl anti-fade GLOX buffer (minus enzymes). Leave it to equilibrate for 1-2 min.
- Remove anti-fade GLOX buffer (minus enzymes) and add 100 μl of anti-fade GLOX buffer (containing enzymes) to each slide.
- Cover the samples with a coverslip, remove excess anti-fade GLOX buffer (plus enzymes) and seal with nail varnish.
- Image the slides the same day to avoid sample drying and fluorophore fading.
- Remove the cover slip and wash the samples 2 times in 200 μl wash buffer for 5 min on the slide.
- Imaging
- Images are acquired on a wide-field epifluorescence microscope, with an optical sectioning size of 0.2 μm per z-plane and spanning the entire volume of the cell (around 5 μm, depending on cell size). Exposure times will vary depending on your microscope setup (light source, objective, etc.). We acquired our images using a Zeiss Elyra PS with a 100x oil-immersion objective (1.46 NA) and a cooled electron multiplying-CCD (charge-coupled device) Andor iXon 897 camera (512 x 512, QE > 90%). For probes labeled with Quasar 570 an excitation line of 561 nm was used and signals were detected at 570-640 nm; for probes labelled with Quasar 670 an excitation line of 642 nm and signals were detected at 655-710 nm; for DAPI an excitation line of 405 nm and signals were detected at wavelengths of 420-480 nm. Typically, we used 30 msec exposure time for DAPI, and 200-300 msec for Quasar dyes.
- After image acquisition, Z-stacks can be reduced to a 2D dataset using the maximum projection command in ImageJ (Image/Stack/Z Project). For a detailed description of this step please refer to ImageJ User Guide available at: https://imagej.nih.gov/ij/docs/guide/user-guide.pdf. The maximum projection command will result in an image with diffraction-limited spots corresponding to single RNAs. When exonic probes are used in conjunction with intronic probes for the same gene, some spots will co-localize within nuclei, corresponding to nascent RNA molecules that were being transcribed at the time of fixation (Figure 4).
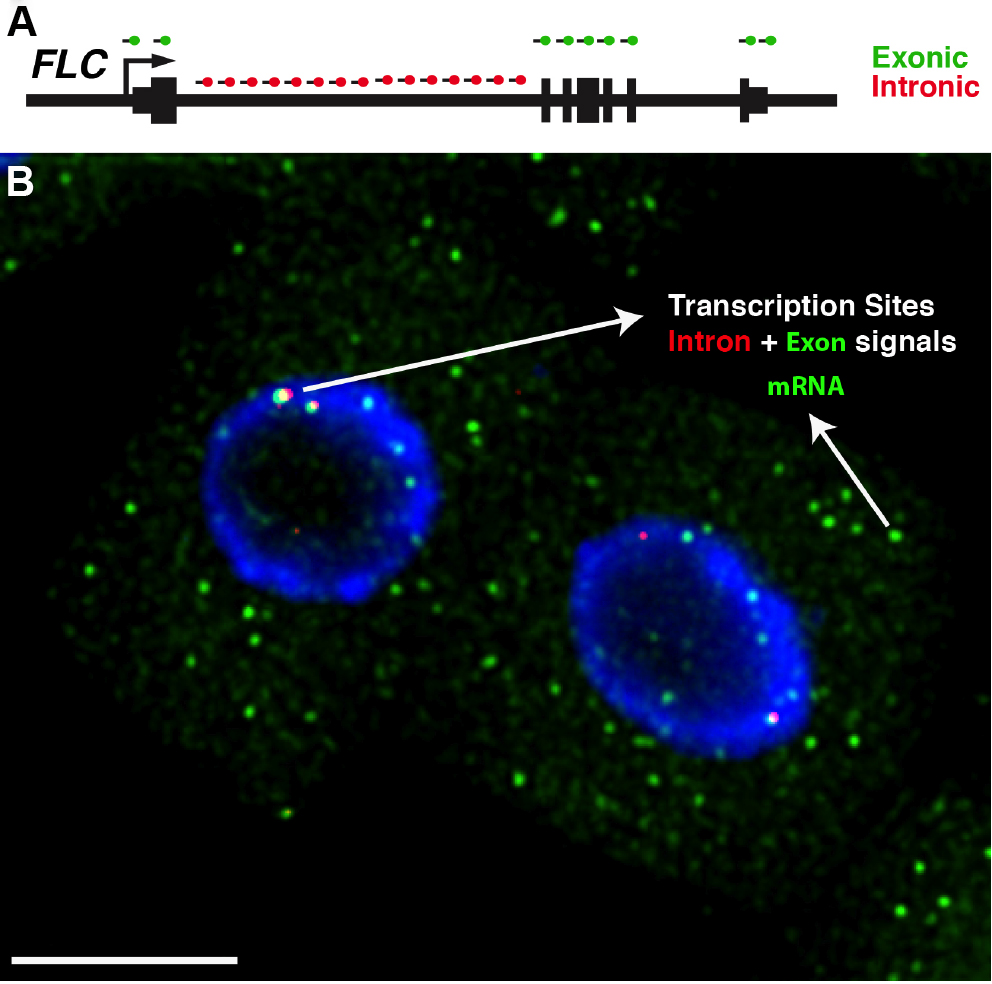
Figure 4. Detection of FLC transcripts in single cells. A. Schematic of the probes used to detect FLC transcripts: intronic (red) and exonic (green). FLC exons are represented as thick lines and introns shown as thin lines. B. Representative image showing FLC mRNA (green) and FLC nascent RNA (red) expression in Arabidopsis thaliana root cells. The diffraction-limited spots in the cytoplasm correspond to single FLC mRNA molecules (green), and the intense spots co-localizing with intronic signals (red) correspond to transcription sites. Depending on the cell cycle stage one can detect up to 2 dots (G1 phase) or 4 dots (G2 phase) for the intronic signals. The nucleus is stained with DAPI and shown in blue. Scale bar = 5 μm.
- Images are acquired on a wide-field epifluorescence microscope, with an optical sectioning size of 0.2 μm per z-plane and spanning the entire volume of the cell (around 5 μm, depending on cell size). Exposure times will vary depending on your microscope setup (light source, objective, etc.). We acquired our images using a Zeiss Elyra PS with a 100x oil-immersion objective (1.46 NA) and a cooled electron multiplying-CCD (charge-coupled device) Andor iXon 897 camera (512 x 512, QE > 90%). For probes labeled with Quasar 570 an excitation line of 561 nm was used and signals were detected at 570-640 nm; for probes labelled with Quasar 670 an excitation line of 642 nm and signals were detected at 655-710 nm; for DAPI an excitation line of 405 nm and signals were detected at wavelengths of 420-480 nm. Typically, we used 30 msec exposure time for DAPI, and 200-300 msec for Quasar dyes.
Data analysis
- For our image analysis we used a publically available mRNA counting programme that had been optiimised for our experimental setup. Details about how to install this programme on a Mac computer can be found at https://github.com/JIC-CSB/FISHcount.
Note: We recommend FISH-quant for researchers wishing to quantify FISH data on Windows machines (Mueller et al., 2013). - Figure 6 shows examples of ‘RNA identification’, ‘Cell Segmentation’ and ‘mRNA Per Cell’ output images that are generated by FISHcount. After performing analysis to a whole dataset, we recommend manually inspecting each ‘mRNA Per Cell’ output image (Figure 6D) to ensure that the image analysis workflow has not generated inaccurate results through incorrect segmentation. A detailed explanation of the image analysis workflow can also be found the Methods section in Duncan et al., 2016 (Figure 5).
- Data can then recorded and plotted as shown in Figure 7 using GraphPad Prism or MS Excel.
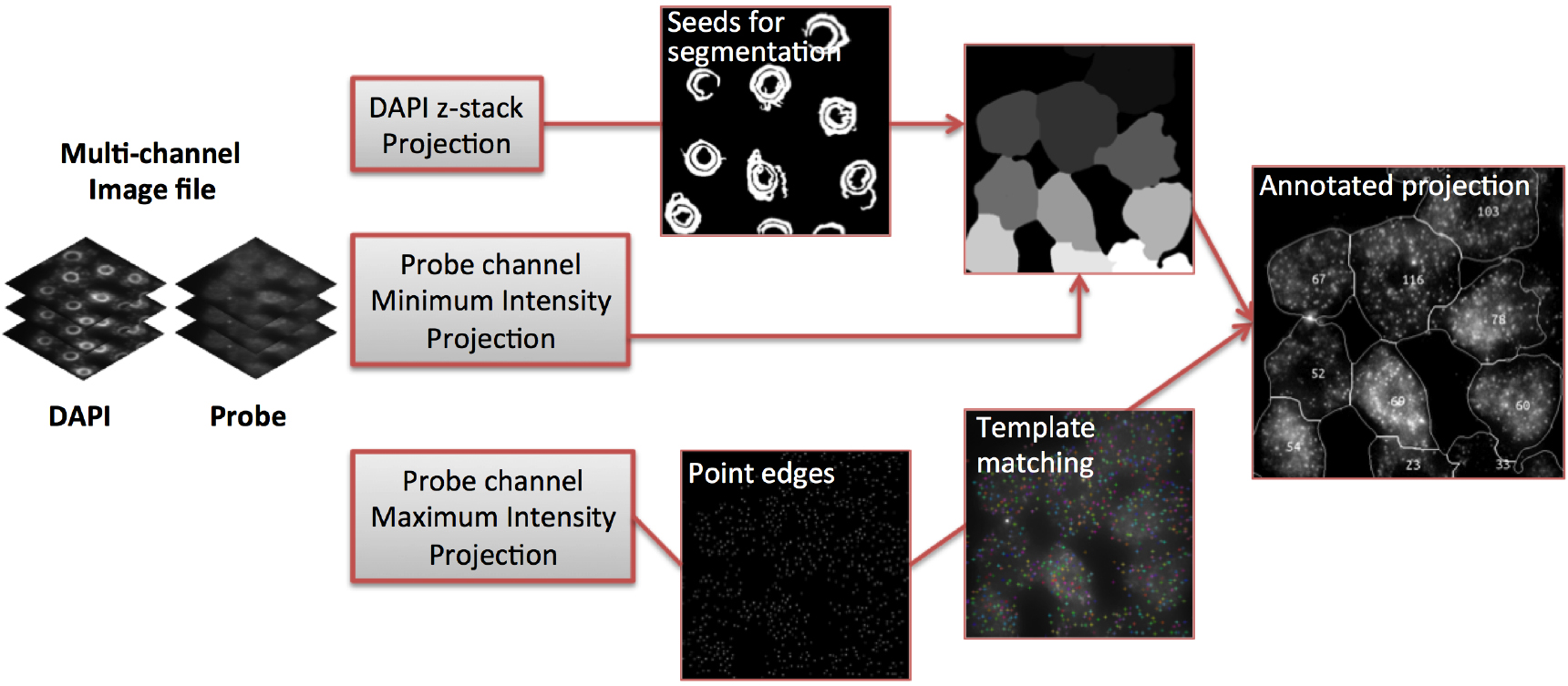
Figure 5. Image analysis workflow (adapted from Duncan et al., 2016)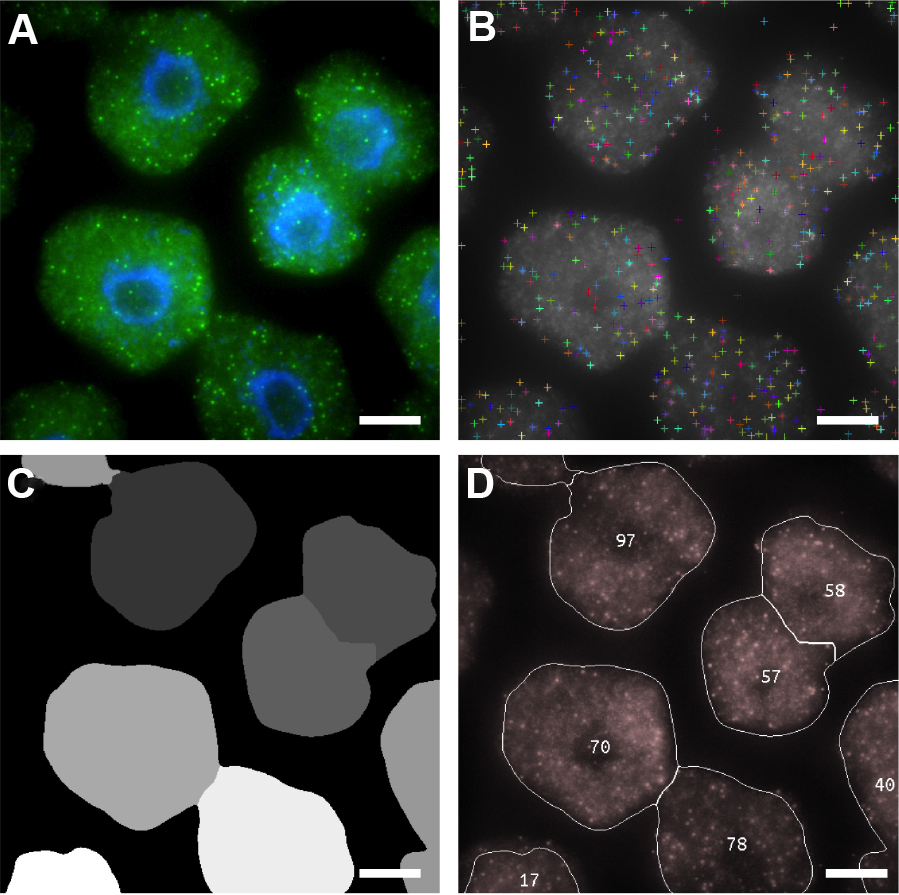
Figure 6. Automated image analysis of FLC mRNA. A. Representative maximum projection image of cells labeled with FLC mRNA probes (green). DNA labeled with DAPI (blue). B-D. Screen shots showing sequential detection steps used to determine positive mRNA signals. B. FLC mRNA spot locations; C. Cell Segmentation; D. Output image with number of mRNAs per cell. Scale bars = 5 μm.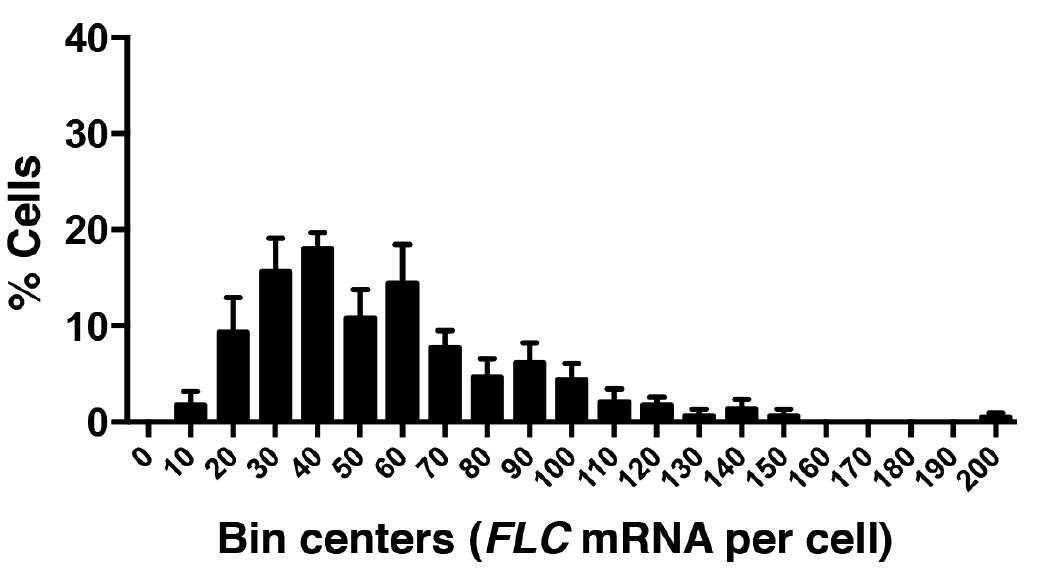
Figure 7. Frequency distribution of FLC mRNA molecules per cell. A total of 520 cells were analyzed from three experiments. Error bars are ± SEM.
Recipes
- Murashige and Skoog medium
0.025 mg/L CoCl2·6H2O
0.025 mg/L CuSO4·5H2O
36.7 mg/L Na·Fe-EDTA
6.2 mg/L H3BO3
0.83 mg/L KI
16.9 mg/L MnSO4·2H2O
0.25 mg/L Na2MoO4·2H2O
8.6 mg/L ZnSO4·7H2O
332.02 mg/L CaCl2·2H2O
170 mg/L KH2PO4
1,900 mg/L KNO3
180.5 mg/L MgSO4·7H2O
1,650 mg/L NH4NO3 (pH 5.8)
1% sucrose
0.5% phytagel - Wash buffer
10% formamide
2x SSC - Hybridization solution
100 mg/ml dextran sulphate
10% formamide
2x SSC - smFISH probe stock (25 μM)
Stellaris 5 nmol oligo probe set
200 μl Tris-EDTA buffer solution - DAPI solution
100 ng/μl DAPI
10% formamide
2x SSC - Anti-fade GLOX buffer (minus enzymes)
0.4% glucose
10 nM Tris-HCl
2x SSC - Anti-fade GLOX buffer (containing enzymes)
For a final volume of 102 μl, mix:
100 μl anti-fade GLOX minus enzyme solution
1 μl glucose oxidase
1 μl bovine liver catalase suspension (mildy vortexed)
Acknowledgments
This work was supported by the UK Biotechnology and Biological Sciences Research Council (BBSRC) grant BB/K00008X/1 and the Earth and Life Systems Alliance (a collaborative venture between John Innes Centre and University of East Anglia). S.R. acknowledges support from 3.3-GRO/1162118STP from Humboldt Foundation (Germany). S.D. acknowledges support from OpenPlant Grant BB/L014130/1. C.D. acknowledges support from European Research Council Advanced grant MEXTIM and BBSRC Institute Strategic Programme grant BB/J004588/1. The original work was published in Duncan et al. (2016) and Rosa et al. (2016).
References
- Duncan, S., Olsson, T. S. G., Hartley, M., Dean, C. and Rosa, S. (2016). A method to detect single molecules of RNA in Arabidopsis thaliana. Plant Methods 12(1): 1-10
- Mueller, F., Senecal, A., Tantale, K., Marie-Nelly, H., Ly, N., Collin, O., Basyuk, E., Bertrand, E., Darzacq, X. and Zimmer, C. (2013). FISH-quant: automatic counting of transcripts in 3D FISH images. Nat Methods 10(4): 277-278.
- Raj, A., van den Bogaard, P., Rifkin, S. A., van Oudenaarden, A. and Tyagi, S. (2008). Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods 5(10): 877-879.
- Rosa, S., Duncan, S. and Dean, C. (2016). Mutually exclusive sense – antisense transcription at FLC facilitates environmentally induced gene repression. Nat Commun 7: 13031.
- Schindelin, J., Rueden, C. T., Hiner, M. C. and Eliceiri, K. W. (2015). The ImageJ ecosystem: An open platform for biomedical image analysis. Mol Reprod Dev 82(7-8): 518-29.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Duncan, S., Olsson, T. S. G., Hartley, M., Dean, C. and Rosa, S. (2017). Single Molecule RNA FISH in Arabidopsis Root Cells. Bio-protocol 7(8): e2240. DOI: 10.21769/BioProtoc.2240.
Category
Plant Science > Plant molecular biology > RNA > RNA detection
Molecular Biology > RNA > RNA detection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









