- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Isolation of Mononuclear Cell Populations from Ovarian Carcinoma Ascites
Published: Vol 7, Iss 7, Apr 5, 2017 DOI: 10.21769/BioProtoc.2219 Views: 11735
Reviewed by: Lee-Hwa TaiJalaj GuptaRalph Bottcher

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Selective Enrichment and Identification of Cerebrospinal Fluid-Contacting Neurons In Vitro via PKD2L1 Promoter-Driven Lentiviral System
Wei Tan [...] Qing Li
Nov 20, 2025 1343 Views

Revisiting Primary Microglia Isolation Protocol: An Improved Method for Microglia Extraction
Jianwei Li [...] Guohui Lu
Dec 5, 2025 1475 Views

FLARE: A Flow Cytometry–Based Fluorescent Assay for Measuring HSV-1 Nuclear Egress
Bing Dai [...] Ekaterina E. Heldwein
Jan 5, 2026 594 Views
Abstract
Ovarian cancer is one of the most fatal tumors in women. Due to a lack of symptoms and adequate screening methods, patients are diagnosed at advanced stages with extensive tumor burden (Jelovac and Armstrong, 2011). Interestingly, ovarian cancer metastasis is generally found within the peritoneal cavity rather than other tissues (Lengyel, 2010; Tan et al., 2006). The reason behind this tissue tropism of the peritoneal cavity remains elusive. A prominent feature of this selectivity is ascites, the accumulation of fluid within the peritoneal cavity, containing, amongst others, immune cells, tumor cells and various soluble factors that can be involved in the progression of ovarian cancer (Kipps et al., 2013). The protocol described here is used to isolate mononuclear cells from ascites to study the functionality of the immune system within the peritoneal cavity.
Keywords: Ovarian cancerBackground
Gradient centrifugation using LymphoprepTM is a standard protocol to isolate peripheral blood mononuclear cells (PBMCs). We slightly adjusted the protocol, regarding the sample preparation and amount of washing steps, in order to isolate mononuclear cells from ascites.
Materials and Reagents
- 50 ml tubes (Greiner Bio One International, catalog number: 227261 )
- Cell strainer 100 μm (Corning, Falcon®, catalog number: 352360 )
- 5 ml pipette (VWR, catalog number: VWR612-3702 )
- 10 ml pipette (VWR, catalog number: VWR612-3700 )
- 25 ml pipette (VWR, catalog number: VWR612-3697 )
- 225 ml tubes (Corning, Falcon®, catalog number: 352075 )
- 5 ml polypropylene round-bottom tube (Corning, Falcon®, catalog number: 352063 )
- Pre-separation filter 30 μm (Miltenyi Biotec, catalog number: 130-041-407 )
- Türk’s solution (EMD Millipore, catalog number: 1092770100 )
- LymphoprepTM (Alere Technologies, Axis-Shield, catalog number: 1114547 )
- Trypan blue stain (Thermo Fisher Scientific, GibcoTM, catalog number: 15250061 )
- FcR blocking reagent (Miltenyi Biotec, catalog number: 120-000-442 )
- Antibodies
- Anti-CD45-V450 (BD, BD Biosciences, catalog number: 560367 )
- Anti-CD19-FITC (Dako Cytomation, catalog number: F0768 )
- Anti-CD20-FITC (BD, BD Biosciences, catalog number: 345792 )
- Anti-CD56-FITC (BD, BD Biosciences, catalog number: 345811 )
- Anti-CD1c-PE (BDCA1) (Miltenyi Biotec, catalog number: 130-090-508 )
- Anti-CD14-PerCP (BD, BD Biosciences, catalog number: 345786 )
- Anti-HLA-DR-PE-Cy7 (BD, BD Biosciences, catalog number: 335830 )
- Anti-CD4-APC-Cy7 (BD, BD Biosciences, catalog number: 557871 )
- Anti-CD3-BV510 (BD, BD Biosciences, catalog number: 563109 )
- Ethylenediaminetetraacetic acid (EDTA) (EMD Millipore, catalog number: 1084211000 )
- Bovine serum albumin (BSA) (Roche Diagnostics, catalog number: 10735108001 )
- Phosphate-buffered saline (PBS) (Braun Melsungen, catalog number: 362 3140 )
- X-VIVO 15 (Lonza, catalog number: BE02-060Q )
- Antibiotic/Antimycotic (AA) (Thermo Fisher Scientific, GibcoTM, catalog number: 15240062 )
- Human serum (HS) (Bloodbank Rivierenland)
- 0.5 M EDTA (see Recipes)
- 10% BSA (see Recipes)
- Diluting buffer (see Recipes)
- Wash buffer (see Recipes)
- Staining buffer (see Recipes)
- Medium (see Recipes)
Equipment
- Centrifuge (Hettich Instruments, model: Rotanta 460R )
- Cell sorter (BD, model: FACS Aria II)
- Magnetic stirrer (Heidolph instruments, model: MR Hei-Mix S )
Procedure
- Isolation of immune cells from ascites
- Take four 50 ml tubes and put a 100 μm cell strainer on top of each of them.
- Pipette the ascites on top of the cell strainer and collect the ascites in the 50 ml tube underneath. Transfer the filtered ascites to a 225 ml Falcon tube.
Note: Filtering the ascites removes tissue, fat or blood clumps that could interfere with cell isolation and cell sorting. - Centrifuge at 4 °C:
Maximum acceleration
Running time 15 min at 490 x g
Maximum break - Pour away the supernatant. Be careful, as the pellet detaches easily when high amounts of red blood cells are present.
- Resuspend the pellet in diluting solution and count the cells with Türk’s solution (use Türks:cells in a ratio of 10:1). Dilute the cell suspension to a concentration of 100 x 106 cells per 10 ml in diluting buffer. Add 10 ml of the cell suspension per 50 ml tube and complete to 30 ml with diluting solution.
Note: Do not put more than 100 x 106 ascites-derived cells on top of the LymphoprepTM layer, as this may interfere with the separation of the cells. - Pipette 10 ml LymphoprepTM underneath the diluted cells (Figures 1A-1C). Take up 12 ml LymphoprepTM using a 10 ml pipette. Let the pipette tip slide carefully along the side of the tube until it reaches the bottom. Detach the pipette from the pipette boy and wait for the LymphoprepTM to leak out. Pull out the pipette until the tip reaches the top of the LymphoprepTM layer. Put your finger on top of the pipette to close the system and remove the pipette from the tube. There should be approximately 2 ml of LymphoprepTM left in the pipette.
- Centrifuge at room temperature:
Acceleration time 240 sec (R/1)
Running time 20 min at 970 x g
No break - Carefully remove the interphase with a 5 ml pipette and collect it in a new 50 ml tube (Figure 1D). Bring the volume to 50 ml with diluting solution.
Note: Be careful when removing the interphase. Hold the pipette tip slightly above the interphase. This prevents removing the LymphoprepTM solution along with the interphase layer. - Centrifuge at room temperature:
Maximum acceleration
Running time 10 min at 640 x g
Maximum break - Pour away the supernatant and resuspend the pellet in 50 ml washing solution.
- Centrifuge at 4 °C:
Maximum acceleration
Running time 5 min at 490 x g
Maximum break - Pool the cells into one 50 ml tube and repeat washing with washing solution until the supernatant is clear.
Note: It usually takes 1 or 2 washing steps until the supernatant is clear. - Count the cells with trypan blue stain (dilute cells and trypan blue in a ratio of 1:1).
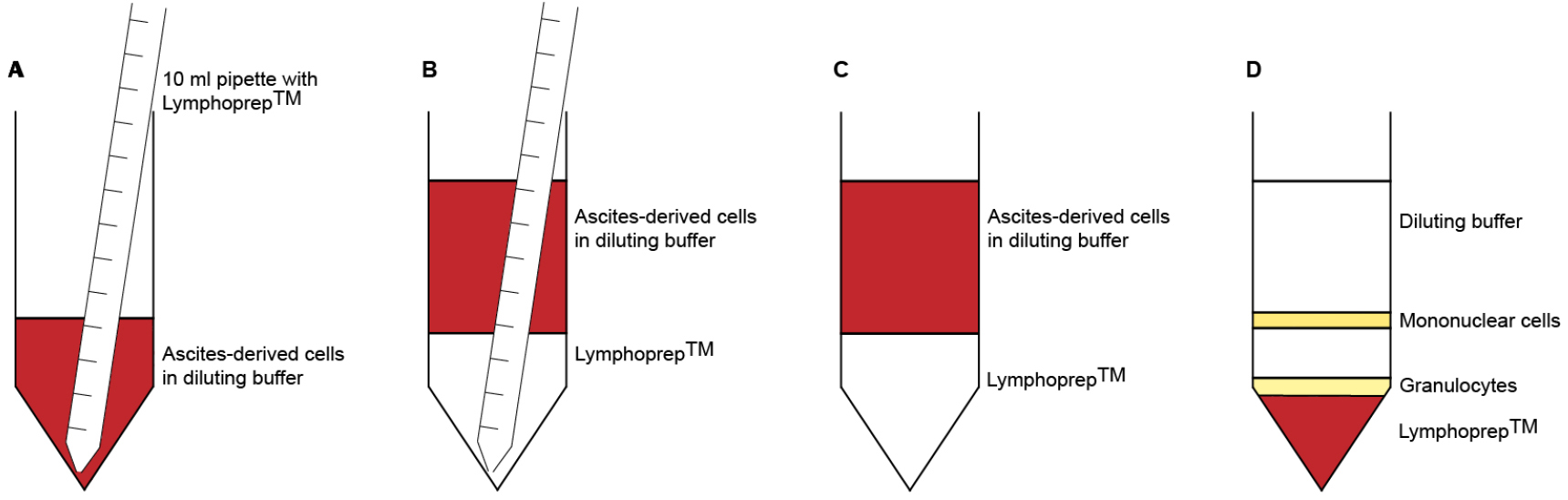
Figure 1. Density gradient centrifugation. A. Use a 10 ml pipette to take up 12 ml LymphoprepTM. Slide the pipette tip along the tube wall until it reaches the bottom. B. Disconnect the pipette from the pipette boy and wait for the LymphorpepTM to leak out. C. Carefully and slowly remove the pipette from the tube. D. After centrifugation, immune cells accumulate in different layers, based on their density. Mononuclear cells (B cells, T cells, NK cells, monocytes) are located in the interphase, between the diluting buffer and the LymphoprepTM. Polymorphonuclear cells (granulocytes) can be found on top of the red blood cells, which settle at the bottom of the tube.
- Take four 50 ml tubes and put a 100 μm cell strainer on top of each of them.
- FACS staining
- Centrifuge cells at 4 °C:
Maximum acceleration
Running time 5 min at 490 x g
Maximum break - Pour away the supernatant and dilute cells with staining buffer to a concentration of 50 x 106 cells/ml.
- Transfer 50 x 106 cells to a new 50 ml tube.
- Centrifuge at 4 °C:
Maximum acceleration
Running time 5 min at 490 x g
Maximum break - Pour away the supernatant. The remaining liquid has a volume of approximately 300 μl.
- Add 50 μl FcR blocking reagent (1 μl per 1 x 106 cells) and incubate for 5 min at 4 °C.
- Add 1 ml staining buffer and centrifuge at 4 °C:
Maximum acceleration
Running time 5 min at 490 x g
Maximum break - Add 12.5 μl of anti-HLA-DR-PE-Cy7 (0.25 μl antibody per 1 x 106 cells) and 25 μl of each of the remaining antibodies (0.5 μl antibody per 1 x 106 cells): anti-CD45-V450, anti-CD19-FITC, anti-CD20-FITC, anti-CD56-FITC, anti-CD1c-PE, anti-CD14-PerCP, anti-HLA-DR-PE-Cy7, anti-CD4-APC-Cy7, anti-CD3-BV510.
- Incubate for 20 min at 4 °C and in the dark.
- Add 10 ml staining buffer and centrifuge at 4 °C:
Maximum acceleration
Running time 5 min at 490 x g
Maximum break - Pour away the supernatant and repeat step B10 two more times.
- Resuspend cells in 1 ml staining buffer.
Note: For cell sorting, do not use higher concentrations than 50 x 106 cells/ml, as this will result in too many events per seconds that are measured by the FACS Aria II and interferes with proper sorting. - Place a 30 μm filter on top of a new 5 ml polypropylene round-bottom tube and pass the cells through the filter into the new tube.
Note: Passing the cells through a 30 μm filter removes cell clumps that could obstruct the nozzle of the cell sorter. - Prepare three 5 ml polypropylene round-bottom tubes with 1 ml medium to collect the cells in after sorting.
- Centrifuge cells at 4 °C:
- Cell sorting
- Cell sorting is performed at room temperature. The collection tubes within the sorter are kept at 4 °C.
Note: Keep the cells before and after sorting on ice to increase cell viability. - Select the 85 μm nozzle on the FACS Aria II, to reduce the pressure on sorted cells.
- The following gating strategy was used to sort the different cell populations (Figure 2).
- Collect the cells in a 5 ml polypropylene round-bottom tube with 1 ml medium.
Note: Shake the round-bottom tube before collecting the cells to cover the surface of the tube with medium. This prevents the cells from sticking to the tube wall.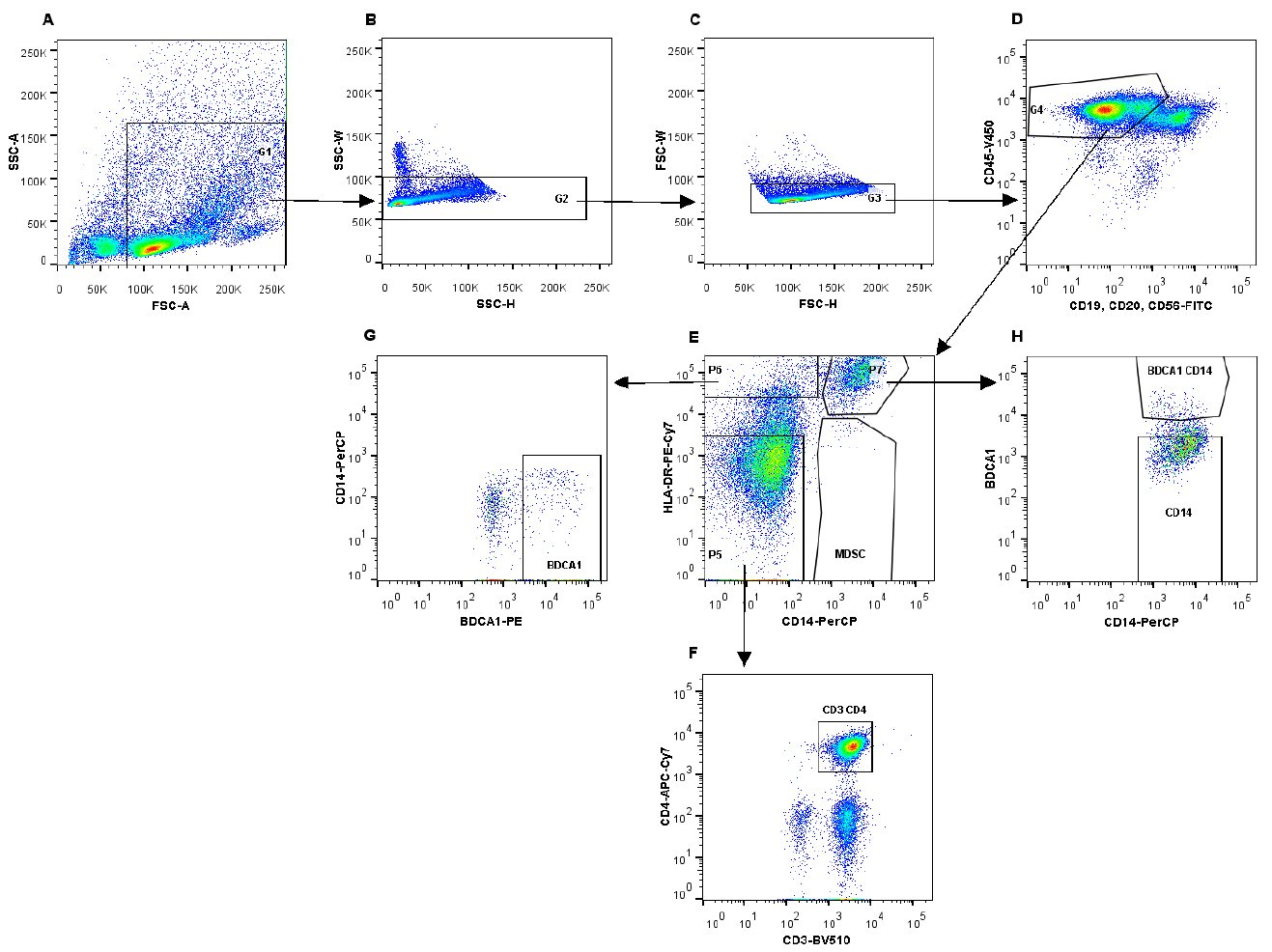
Figure 2. Gating strategy to isolate immune cell subsets using fluorescent activated cell sorting. A. Gating on immune cells on the FSC/SSC plot; B and C. Selection of single cells; D. Gating on immune cell population, which is CD45-V450 positive (G4), thereby excluding B cells and NK cells. E. Different immune cell populations can be identified, based on the expression of HLA-DR-PE-Cy7 and CD14-PerCP. Myeloid-derived suppressor cells are CD14+HLA-DR-. F. From gate G5, CD3+CD4+ T cells were identified and isolated. G. Gate G6 (HLA-DR+CD14-) was used to identify BDCA1+ cells. H. CD14+HLA-DR+ cells (P2) can be further subdivided into CD14+ and BDCA1+CD14+ cells.
- Cell sorting is performed at room temperature. The collection tubes within the sorter are kept at 4 °C.
Data analysis
The isolated immune cell subsets from ascites can be used for different experiments, like mixed lymphocyte reaction, suppressor assays or antigen uptake assays, as published in Bakdash et al., 2016.
Notes
- As large amounts of ascites can be collected, it is important to spin it down first in order to enrich the cells and dissolve them in a smaller volume.
- LymphoprepTM must be protected from light and stored at room temperature to ensure good separation.
- Keep diluting solution and EDTA at RT.
- 10% BSA and washing solution need to be stored at 4 °C.
- When preparing 0.5 M EDTA it is important to note that the EDTA only dissolves at a pH of 8.
- FITC is used as a dump channel. The three FITC labeled antibodies CD19, CD20 and CD56 are used to exclude B cells and NK cells.
Recipes
Note: All buffers and dilutions are stable when stored at the recommended temperature and used under sterile conditions.
- 0.5 M EDTA (store at RT)
186.1 g EDTA
800 ml dH2O
Stir vigorously on a magnetic stirrer and adjust pH to 8.0
Bring volume to 1 L with dH2O - 10% BSA (store at 4 °C)
10 g BSA
100 ml PBS - Diluting buffer (store at RT)
5 ml 0.5 M EDTA
500 ml PBS - Wash buffer (store at 4 °C)
5 ml 0.5 M EDTA
5 ml 10% BSA
500 ml PBS - Staining buffer (store at 4 °C)
PBS
1% BSA
1% EDTA - Medium (store at 4 °C)
100 ml X-VIVO 15
1 ml AA
2 ml HS
Acknowledgments
This work was supported by grant 951.03.002 from the Netherlands Organization for Scientific Research (NWO) and the Netherlands Institute of Regenerative Medicine (NIRM, FES0908). IJMdV is recipient of NWO Vici grant 918.14.655.
The procedure for the isolation of mononuclear cells from ascites was adapted from the standard protocol used for the isolation of PBMCs (Bakdash et al., 2016).
References
- Bakdash, G., Buschow, S. I., Gorris, M. A., Halilovic, A., Hato, S. V., Skold, A. E., Schreibelt, G., Sittig, S. P., Torensma, R., Duiveman-de Boer, T., Schroder, C., Smits, E. L, Figdor, C. G., de Vries, I. J. (2016). Expansion of a BDCA1+CD14+ myeloid cell population in melanoma patients may attenuate the efficacy of dendritic cell vaccines. Cancer Res 76(15): 4332-4346.
- Jelovac, D. and Armstrong, D. K. (2011). Recent progress in the diagnosis and treatment of ovarian cancer. CA Cancer J Clin 61(3): 183-203.
- Kipps, E., Tan, D. S. and Kaye, S. B. (2013). Meeting the challenge of ascites in ovarian cancer: new avenues for therapy and research. Nat Rev Cancer 13(4): 273-282.
- Lengyel, E. (2010). Ovarian cancer development and metastasis. Am J Pathol 177(3): 1053-1064.
- Tan, D. S., Agarwal, R. and Kaye, S. B. (2006). Mechanisms of transcoelomic metastasis in ovarian cancer. Lancet Oncol 7(11): 925-934.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Wefers, C., Bakdash, G., Moreno Martin, M., Duiveman-de Boer, T., Torensma, R., Massuger, L. F. and de Vries, I. J. M. (2017). Isolation of Mononuclear Cell Populations from Ovarian Carcinoma Ascites. Bio-protocol 7(7): e2219. DOI: 10.21769/BioProtoc.2219.
Category
Cell Biology > Cell isolation and culture > Cell isolation
Cell Biology > Cell-based analysis > Flow cytometry
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












