- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Purification of N-coronafacoyl Phytotoxins from Streptomyces scabies
Published: Vol 7, Iss 7, Apr 5, 2017 DOI: 10.21769/BioProtoc.2214 Views: 8446
Reviewed by: Valentine V TrotterRosario Gomez-GarciaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

β-lactamase (Bla) Reporter-based System to Study Flagellar Type 3 Secretion in Salmonella
Fabienne F. V. Chevance and Kelly T. Hughes
Jun 20, 2023 1776 Views

Determination of Poly(3-hydroxybutyrate) Content in Cyanobacterium Synechocystis sp. PCC 6803 Using Acid Hydrolysis Followed by High-performance Liquid Chromatography
Janine Kaewbai-ngam [...] Tanakarn Monshupanee
Aug 20, 2023 1828 Views

An HPLC-based Assay to Study the Activity of Cyclic Diadenosine Monophosphate (C-di-AMP) Synthase DisA from Mycobacterium smegmatis
Avisek Mahapa [...] Dipankar Chatterji
Dec 20, 2024 1791 Views
Abstract
This procedure is used for large-scale purification of N-coronafacoyl phytotoxins that are produced by the potato common scab pathogen Streptomyces scabies. The procedure employs organic extraction of S. scabies culture supernatants under alternating basic and acidic conditions in order to preferentially isolate the phytotoxin - containing carboxylic acid fraction of the supernatant. Preparative thin layer chromatography and semi-preparative reverse phase - high performance liquid chromatography are then used to further purify the individual N-coronafacoyl phytotoxins of interest.
Keywords: StreptomycesBackground
Potato common scab is an economically important crop disease that is caused by Gram-positive, filamentous, soil bacteria from the genus Streptomyces. The first described and best characterized scab - causing Streptomyces spp. is Streptomyces scabies (syn. S. scabiei), which has a worldwide distribution (Bignell et al., 2010). Current control practices for common scab disease management include crop rotation, irrigation and soil fumigation; however, these strategies often fail, produce inconsistent results or are environmental unfriendly (Dees and Wanner, 2012). In order to develop better control strategies for the disease, we must first understand the molecular mechanisms used by S. scabies to infect the plant and to induce disease symptoms. Research has shown that the ability of S. scabies to cause disease is due to the production of virulence factors that play different roles during the infection process. Among the known or potential virulence factors that are produced by S. scabies is a family of plant toxins referred to as the N-coronafacoyl phytotoxins (also known as the COR-like metabolites), which resemble the plant hormone jasmonic acid and may function to suppress the plant immune response during pathogen infection (Bignell et al., 2010; Fyans et al., 2015). The primary coronafacoyl phytotoxin produced by S. scabies is N-coronafacoyl-L-isoleucine (CFA-L-Ile; Figure 1), which consists of the polyketide metabolite coronafacic acid linked via an amide bond to L-isoleucine. In addition, other N-coronafacoyl phytotoxins containing different isoleucine isomers or different amino acids (e.g., valine) can be produced in minor amounts (Fyans et al., 2015; Bown et al., 2016). 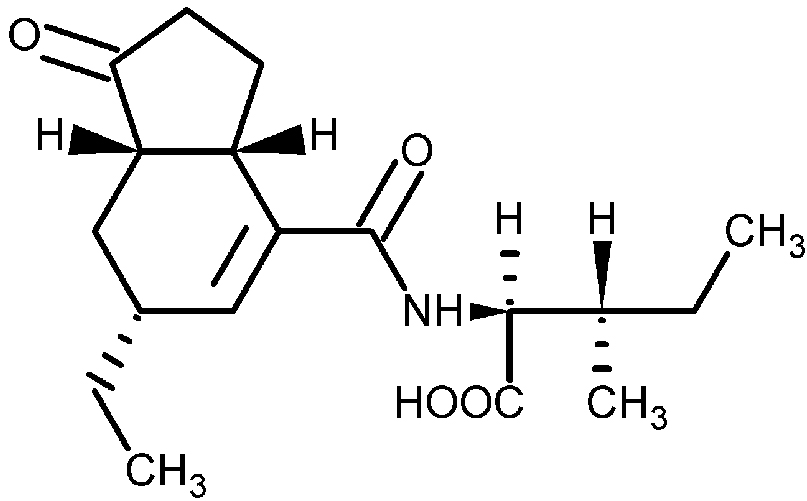
Figure 1. Structure of N-coronafacoyl-L-isoleucine (CFA-L-Ile) produced by Streptomyces scabies
The protocol described here was developed to isolate and purify N-coronafacoyl phytotoxins and their biosynthetic intermediates from large-scale cultures of S. scabies for purposes of structural and functional characterization. Previously, Fyans et al. (2015) described a protocol that was based in part on a published procedure for the isolation of the related N-coronafacoyl phytotoxin coronatine (COR) from cultures of the Gram-negative plant pathogenic bacterium Pseudomonas syringae (Palmer and Bender, 1993). As outlined by Fyans et al. (2015), strains of S. scabies are cultured in a soy flour mannitol broth (SFMB) medium, which promotes the production of the coronafacoyl phytotoxins, and then the culture supernatants are subjected to a two-step extraction with organic solvent under basic and acidic conditions in order to selectively isolate the phytotoxin - containing carboxylic acid fraction of the culture supernatants. The phytotoxins are then further purified using a combination of preparative thin layer chromatography (TLC) and semi-preparative reverse phase - high performance liquid chromatography (RP - HPLC). More recently, we described a modified version of this protocol in which we incorporated additional extraction steps using an aqueous solution of potassium bicarbonate (Bown et al., 2016). This modification was based on the procedure described by Mitchell and Frey (1986) for the isolation of P. syringae N-coronafacoyl phytotoxins, and we found that the incorporation of the additional extraction steps significantly improved the purity of the final S. scabies phytotoxin preparations. Moreover, we modified the organic solvent for the N-coronafacoyl phytotoxins by the addition of a small amount of acid, which significantly improved the solubility and yield of the purified phytotoxins for downstream structural and functional studies.
Here, we present the detailed step-by-step protocol for how we currently purify the S. scabies N-coronafacoyl phytotoxins in our laboratory.
Materials and Reagents
- pH test strips (VWR, BDH®, catalog number: BDH35309.606 )
- Hydrophilic polypropylene membrane filters, 47 mm diameter, 0.45 μm pore size (Pall, catalog number: 66548 )
- FisherbrandTM class B clear glass threaded vials with closures attached, 3.7 ml (Fisher Scientific, catalog number: 03-338A )
- Slip tip syringes, 1 ml (BD, catalog number: 309659 )
- PTFE membrane filters, 0.2 μm pore size, 6 mm diameter (VWR, catalog number: 28145-491 )
- WhatmanTM filter discs, 12.5 cm (Sigma-Aldrich, catalog number: WHA1113125 )
- Conical centrifuge tubes, 50 ml (Corning, Falcon®, catalog number: 352098 )
- Silica gel GF preparative TLC plates with pre-adsorbent zone, 20 x 20 cm, 1,000 μm (Analtech/iChromatography, catalog number: P32013 )
- DMSO mycelial freezer stock of S. scabies (Fyans et al., 2015)
- BactoTM tryptic soy broth medium (BD, BactoTM, catalog number: 211825 )
- Sodium hydroxide (NaOH) (Fisher Scientific, catalog number: BP359-212 )
- Chloroform, ACS grade (VWR, BDH®, catalog number: BDH1109 )
- Hydrochloric acid (HCl), ACS grade (Avantor® Performance Materials, catalog number: 638801 )
- Sodium sulfate anhydrous (Na2SO4), ACS grade (Fisher Scientific, catalog number: S421-500 )
- Ethylene glycol (VWR, BDH®, catalog number: BDH1125 )
- Potassium bicarbonate (KHCO3), ACS grade (Sigma-Aldrich, catalog number: 237205 )
- Methanol (MeOH), HPLC grade (Sigma-Aldrich, catalog number: 34860 )
- Formic acid, reagent grade (Sigma-Aldrich, catalog number: F0507 )
- HiPerSolv CHROMANOR® Acetonitrile (ACN) for HPLC (VWR, BDH®, catalog number: BDH83639.400 )
- Soy flour, defatted (MP Biomedicals, catalog number: 960024 )
- D-mannitol (AMRESCO, catalog number: 0122 )
- Ethyl acetate, ACS grade (Sigma-Aldrich, catalog number: 319902 )
- 2-propanol, HPLC grade (Fisher Scientific, catalog number: A451SK-4 )
- Acetic acid, ACS grade (Sigma-Aldrich, catalog number: 695092 )
- Water, HPLC grade (EMD Millipore, catalog number: WX0008-1 )
- Soy flour mannitol broth (SFMB) medium (see Recipes)
- Preparative TLC mobile phase (see Recipes)
Equipment
- Glass Erlenmeyer flask, 125 ml (Corning, PYREX®, catalog number: C4980125 )
- Glass Erlenmeyer flask, 4 L (Corning, PYREX®, catalog number: C49804L )
- Glass filter holder assembly with funnel, fritted base, stopper and clamp, 47 mm (EMD Millipore, catalog number: XX1004700 )
- NalgeneTM PPCO centrifuge bottles with sealing closure, 250 ml (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 31410250 )
- SorvalTM ST 16R benchtop refrigerated centrifuge (Thermo Fisher Scientific, Thermo ScientificTM, model: SorvalTM ST 16R , catalog number: BCT25)
- Innova® 42R refrigerated incubator shaker, orbit diameter 1.9 cm (Eppendorf, New BrunswickTM, model: Innova® 42R , catalog number: M1335-004)
- 2 L plastic container
- NalgeneTM TeflonTM FEP separatory funnel with closure, 2 L (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 4301-2000 )
- IKA® rotary evaporator system (IKA®, model: RV 10 digital V )
- VWR® refrigerated circulating bath with programmable temperature controller (VWR, model: VWR® Refrigerated Circulating Baths , catalog number: 89202-982)
- KIMAX® Squibb separatory funnel with PTFE stopcock and glass stopper, 125 ml (Kimble Chase Life Science and Research Products, catalog number: 29048F-125 )
- DryFast diaphragm vacuum pump (Welch Vacuum – Gardner Denver, catalog number: 2044 )
- Biohit mLINE® single-channel mechanical pipettor, 2-20 μl (VWR, catalog number: 47745-545 )
- Aldrich® rectangular TLC developing tank (Sigma-Aldrich, model: Z126195 )
- UV lamp with portable cabinet (Analtech/iChromatography, catalog number: A93-04 )
- Metal spatula
- Thermo ScientificTM dry block heater (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 88-860-021 )
- ZORBAX StableBond 80Å C18 semi-preparative HPLC column, 9.4 x 250 mm, 5 μm (Agilent Technologies, catalog number: 880975-202 )
- Chemical safety hood
- KIMAX® graduated filtering flask, 1 L (Kimble Chase Life Science and Research Products, catalog number: 27060-1000 )
- GeneMate vortex mixer (VWR, catalog number: 490000-094 )
- Agilent 1260 Infinity analytical-scale LC purification system with quaternary pump, autosampler, diode array detector and fraction collector (Agilent Technologies, model: 1260 Infinity )
Procedure
- Culture growth
- Prepare a seed culture of S. scabies by inoculating 1 ml of the DMSO freezer stock into 25 ml of tryptic soy broth medium in a 125 ml Erlenmeyer flask, and incubate at 28 °C with shaking (200 RPM) for 48 h.
- Subculture 10 ml of the seed culture into 1 L of SFMB medium in a 4 L Erlenmeyer flask, and incubate at 25 °C with shaking (200 RPM) for 10 days (see Notes 1 and 2).
- Divide the culture into four 250 ml centrifuge bottles and centrifuge at 3,005 x g for 15 min at 4 °C to pellet the cell material. Decant the supernatant into a 2 L plastic container and store at -20 °C until needed. Even if the supernatant is going to be extracted immediately, it is useful to freeze it for a minimum of 24 h (see Note 3).
- Prepare a seed culture of S. scabies by inoculating 1 ml of the DMSO freezer stock into 25 ml of tryptic soy broth medium in a 125 ml Erlenmeyer flask, and incubate at 28 °C with shaking (200 RPM) for 48 h.
- Organic extraction (see Figure 2 and Note 4)
Figure 2. Schematic outline of principle steps performed during the extraction of coronafacoyl phytotoxins from S. scabies culture supernatants- Measure the pH of the culture supernatant using a pH strip, and adjust the pH to 11-12 using a 5 N solution of NaOH (~4-5 ml).
- Decant the supernatant into a 2 L separatory funnel and extract with 0.5 volumes (~450 ml) of chloroform. To perform the extraction, add the chloroform to the separatory funnel, close the funnel stopper, and shake the funnel to mix the contents well. Open the stopcock to release any expressed gas, then repeat the shaking until no more gas is expressed. Allow the organic (bottom) and aqueous (top) layers to separate as much as possible by gravity (~15-20 min; Figure 3A), then open the stopcock and collect the organic layer. If an emulsion layer is present, this can be collected and separated by centrifugation at 3,005 x g for 10 min at 4 °C, and the aqueous layer returned to the separatory funnel (Figure 3B). Discard the organic layer, and repeat the extraction of the aqueous supernatant with another 0.5 volumes of fresh chloroform. Perform the extraction a total of three times, discarding the organic layer after each extraction (see Note 5).
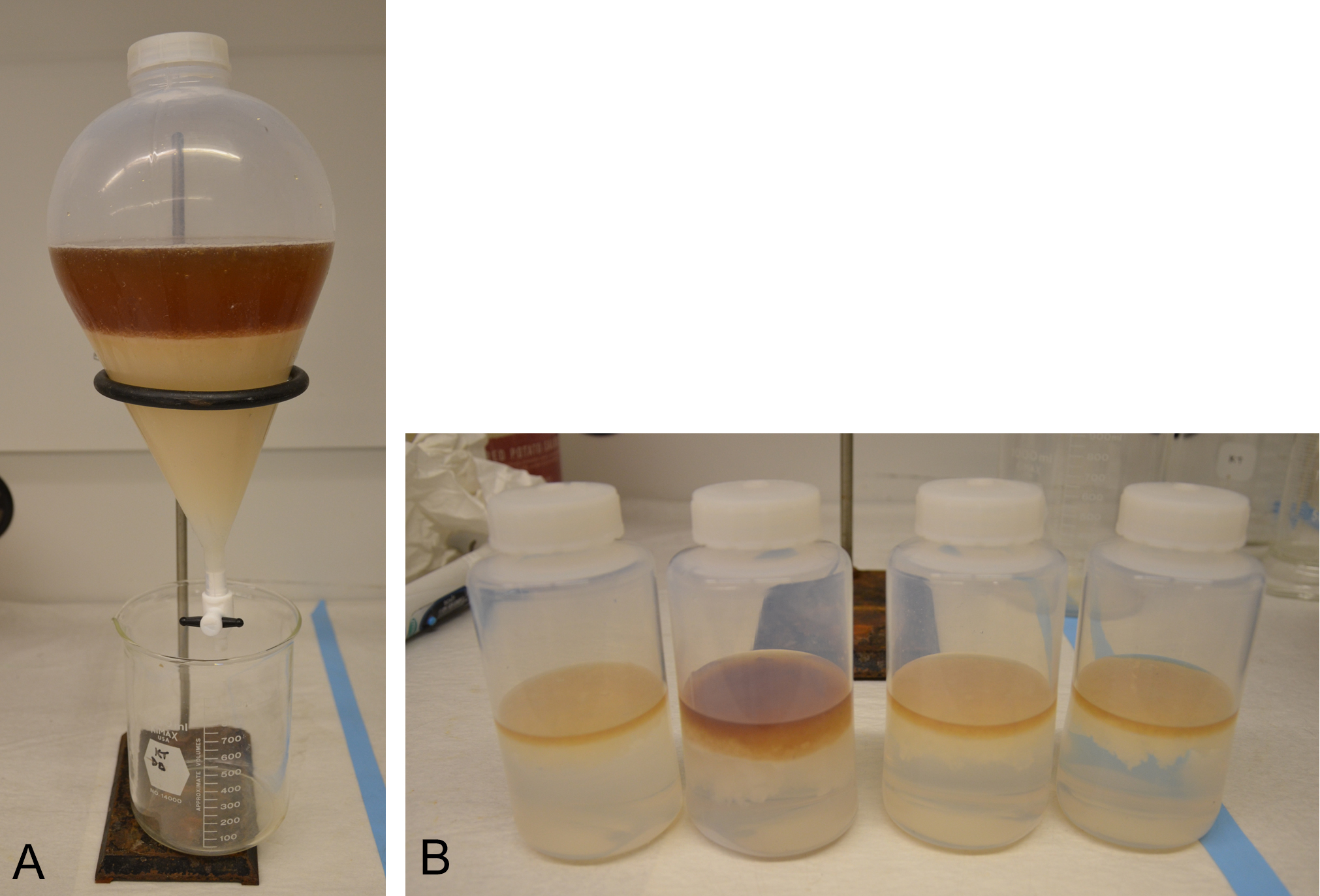
Figure 3. Extraction of S. scabies culture supernatants with chloroform. A. The extraction is performed using a 2 L separatory funnel. After mixing the supernatant and chloroform, the phases are allowed to separate as much as possible by gravity. The upper dark brown layer is the aqueous phase, and the lower layer is the organic phase. B. When an emulsion layer is present, it is collected into centrifuge bottles and the phases are separated by centrifugation. - Acidify the aqueous supernatant to a pH of 1-2 by adding 1 N HCl(aq) (~5 ml).
- Decant the supernatant back into the 2 L separatory funnel and extract with 0.5 volumes (~450 ml) of chloroform. Use the same extraction procedure as described under 2 except that now the organic layer should be collected and saved. Perform the extraction a total of three times, after which the collected organic layers can be pooled and the aqueous layer discarded (see Note 6).
- Remove any residual water from the organic extract by adding ~30 g of anhydrous Na2SO4 and gently swirling the mixture. The anhydrous Na2SO4 is added until it no longer forms clumps, indicating that all of the residual water has been absorbed.
- Filter the organic extract using a 0.45 μm polypropylene membrane filter and a glass filter assembly attached to a vacuum pump. Wash the beaker that contained organic extract as well as the filter assembly with fresh chloroform to recover all of the residual extract.
- Concentrate the organic extract to ~100 ml using a rotary evaporator. The evaporator heating bath (filled with water) should be set to 50 °C, and the condenser should be chilled with a circulating solution of 50% v/v ethylene glycol (chilled to -10 °C using a refrigerated circulating bath).
- Transfer 50 ml of the organic concentrate into each of two 125 ml separatory funnels. Extract each concentrate with 0.5 volumes (25 ml) of a 0.5 M aqueous solution of KHCO3. After mixing, allow ~5 min for the phases to separate by gravity, then collect each phase separately. Repeat the extraction of the organic concentrates two more times, then pool the collected aqueous phases (to give two sets of aqueous extracts, ~75 ml each) and discard the organic extracts (see Note 7).
- Wash the aqueous extracts twice with 0.5 volumes (35-40 ml) of chloroform. To wash, add the chloroform to a separatory funnel containing the aqueous extract, mix, then leave for 5 min to allow the phases to separate. Collect and discard the organic layer, and repeat. After washing, combine the aqueous extracts together (total volume ~140-150 ml).
- Adjust the pH of the aqueous extract to 1-2 using 1 N HCl(aq) (~12-15 ml). The acid will cause a neutralization reaction with the KHCO3 that will produce heat and a large amount of CO2. The acid must be added slowly or the reaction can become violent and will cause the acid to splash. After each 1 ml of acid has been added, gently mix the solution until the CO2 dissipates.
- Pour ~40 ml of the aqueous extract into each of four 125 ml separatory funnels. Extract each aliquot three times with 0.5 volumes (20 ml) of chloroform. Collect and combine all of the organic extracts together, and discard the aqueous layers. Evaporate the organic extract to dryness overnight in an uncovered beaker in a chemical safety hood (see Note 8).
- Measure the pH of the culture supernatant using a pH strip, and adjust the pH to 11-12 using a 5 N solution of NaOH (~4-5 ml).
- Preparative TLC (see Note 4)
- Re-dissolve the dried organic extract in 2-3 ml of MeOH containing 0.1% v/v formic acid by pipetting up and down. Transfer the extract to a clean screw-capped glass vial. Rinse the beaker with an additional 1 ml of MeOH + 0.1% formic acid to recover residual amounts of the extract. Filter the extract into a new screw-capped vial using a 1 ml syringe and a 0.2 μm PTFE membrane filter.
- Pour the TLC mobile phase (see Recipes) into a TLC developing tank and fill to a depth of ~2 cm. Saturate the air in the TLC chamber by placing two pieces of 12.5 cm filter paper into the tank and covering the tank for at least 30 min.
- Mark a 5 x 20 cm analytical TLC plate with a pencil to indicate the submersion point of the mobile phase solution (see Note 9).
- Apply at least ~30-50 μg of a standard of the molecule of interest (or of a comparative analogue; see Note 10) dissolved in 100% MeOH + 0.1% formic acid onto the TLC plate. Apply the standard at least 1 cm from the edge of the plate and at least 1 cm above the submersion mark on the plate.
- Apply ~30-50 μl of the organic extract along ~2 cm of the plate width. Ensure that the applied extract is at least 1 cm from the edge of the plate, 1 cm above the submersion mark on the plate and 1 cm away from the previously applied standard (see Note 11).
- Remove the filter paper discs from the TLC tank and place the TLC plate into the tank and cover. Allow the mobile phase to migrate to within 1 cm of the top of the plate (~20-40 min). Remove the TLC plate and allow to air dry (~15 min). Repeat the process.
- Place the air dried TLC plate under a UV lamp and use a pencil to mark the positions of the applied standard and the N-coronafacoyl phytotoxin of interest in the organic extract (Figure 4).

Figure 4. TLC analysis of the S. scabies organic extract containing CFA-L-Ile. A pure sample (50 µg) of CFA-L-Ile (i) and a small portion (30-50 µl) of the organic extract (ii) containing CFA-L-Ile is spotted onto a 5 x 20 cm silica gel GF analytical TLC plate. Following separation, the plate is visualized under UV light, and the position of the pure standard is marked (blue circle) along with the expected position of the phytotoxin within the extract (blue rectangle). The black arrow indicates the direction of migration of the samples on the plate. - Mark the preparative TLC plate with a pencil to indicate the area of the plate that will be submersed in the mobile phase solution.
- Apply the organic extract along the entire width of the TLC plate, 10 μl at a time. Do not place any sample within 1 cm of either edge of the plate or within 1 cm of the marked submersed area (see Note 11).
- Remove the filter paper discs from the TLC tank and place the TLC plate into the tank and cover. Allow the mobile phase to migrate to within 1 cm of the top of the plate (~1-1.5 h). Remove the plate from the tank and allow to air dry (~20-30 min). Return the plate to the tank and repeat the separation.
- Place the air dried TLC plate under a UV lamp and use a pencil to mark the region of the plate where the N-coronafacoyl phytotoxin migrated to (Figure 5; see Note 12).
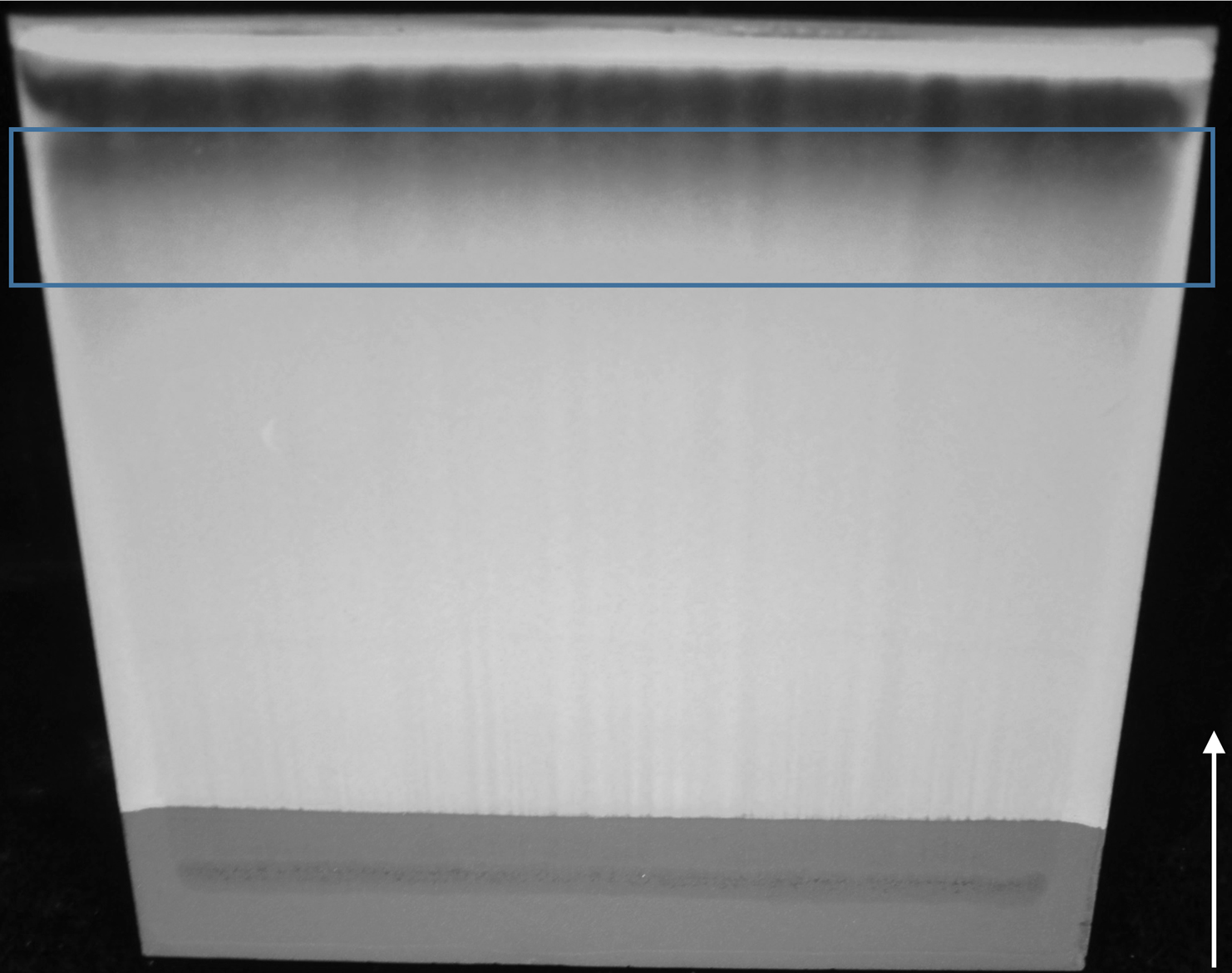
Figure 5. Preparative TLC analysis of the S. scabies organic extract containing CFA-L-Ile. The CFA-L-Ile will be found within the dark smear as indicated by the analytical TLC reference plate (see Figure 4). To ensure complete recovery of the CFA-L-Ile, a large area (indicated by the blue rectangle) of the silica is scraped from the plate. The white arrow indicates the direction of migration of metabolites on the plate. - Scrape the marked region using the flat edge of a metal spatula to remove the silica gel - bound metabolites. Transfer the silica gel to a Falcon conical tube and add 10 ml of MeOH + 0.1% v/v formic acid. Vortex the mixture, and then incubate at room temperature for 1 h, vortexing for 1 min every 10 min.
- Centrifuge the mixture at 3,803 x g for 5 min at room temperature to pellet the silica gel. Transfer the MeOH extract to a clean Falcon tube. Repeat the centrifugation of the silica gel mixture to recover as much of the MeOH extract as possible.
- Filter the MeOH extract using a 1 ml syringe and a 0.2 μm PTFE membrane filter. Concentrate the extract to a final volume of 1-1.5 ml using a dry heating block set to 60 °C (see Note 13).
- Re-dissolve the dried organic extract in 2-3 ml of MeOH containing 0.1% v/v formic acid by pipetting up and down. Transfer the extract to a clean screw-capped glass vial. Rinse the beaker with an additional 1 ml of MeOH + 0.1% formic acid to recover residual amounts of the extract. Filter the extract into a new screw-capped vial using a 1 ml syringe and a 0.2 μm PTFE membrane filter.
- Semi-preparative RP - HPLC
- Begin running the HPLC mobile phase (30% ACN:70% H2O, with 0.1% formic acid) through the C18 semi-preparative HPLC column (held at a constant temperature of 50 °C) at a constant flow rate of 4 ml/min until the column is equilibrated.
- Load 50 μl of the sample onto the column, and use the following mobile phase running conditions for sample separation: hold at 30% ACN:70% H2O for 7.5 min, then increase linearly to 50% ACN:50% H2O over a period of 12.5 min. Hold at 50% ACN:50% H2O for 5 min, then return to initial conditions using a linear gradient over 7.5 min. Hold at the initial conditions for at least 12 min before beginning the next injection (see Note 14).
- Monitor the separated metabolites using a detection wavelength of 230 nm, and collect fractions of the peak that corresponds to the desired N-coronafacoyl phytotoxin (see Note 15).
- Repeat the sample injection, metabolite separation and fraction collection until all of the initial extract has been used.
- Pool the collected fractions for the N-coronafacoyl phytotoxin of interest, and evaporate to dryness using a rotary evaporator. The evaporator heating bath (filled with water) should be set to 70 °C, and the condenser should be chilled with a circulating solution of 50% v/v ethylene glycol (chilled to -10 °C using a refrigerated circulating bath).
- Re-dissolve the pure metabolite in 3 ml of MeOH + 0.1% v/v formic acid. Transfer the solution to a pre-weighed screw cap vial, and dry down in a heating block set to 60 °C.
- Weigh the vial + dried sample, and calculate the weight of the purified sample. Store the dried sample at -20 °C until needed (see Notes 16 and 17).
- Begin running the HPLC mobile phase (30% ACN:70% H2O, with 0.1% formic acid) through the C18 semi-preparative HPLC column (held at a constant temperature of 50 °C) at a constant flow rate of 4 ml/min until the column is equilibrated.
Notes
- We have observed variations in the production levels of the N-coronafacoyl phytotoxins depending on the strain of S. scabies that is used, and therefore the volume of culture that is grown may need to be altered depending on the production efficiency of the particular strain.
- The large-scale cultures can be left to incubate for up to 14 days; however, the levels of metabolite present in the culture will not increase significantly after 10 days.
- The freezing process forces proteins and any other dissolved material out of solution, which will then be removed during the first chloroform extraction. This will make the subsequent extractions cleaner and will allow for faster and more complete phase separation. Use of a plastic container to freeze/thaw the supernatant is recommended, since a glass beaker may crack during the freezing process.
- All steps (with the exception of the centrifugation in steps B2 and C8) should be carried out in a chemical safety hood or a fume hood.
- This step serves as a purification step since many non-carboxylic acid compounds will move into the organic phase under the basic pH conditions used. Carboxylic acids such as the N-coronafacoyl phytotoxins will be in the unprotonated form at the high pH and therefore will remain in the aqueous phase.
- Under the pH conditions used, the N-coronafacoyl phytotoxins are in the pronated form and will therefore be more soluble in the chloroform than in the aqueous culture supernatant.
- The pH of the KHCO3 solution promotes deprotonation of the N-coronafacoyl phytotoxins and their subsequent movement into the aqueous phase.
- This mixing stage produces a large amount of gas. Do not fill the 125 ml funnel with more than 75 ml of aqueous and organic solvent in total. If the funnel contains more than that volume, the expression of gas will force open the stopper or stopcock and the extract will be lost.
- Steps C3-C7 are performed in order to determine the migration point of the N-coronafacoyl phytotoxin of interest. A pure sample of the molecule (~50 μg) and a small portion of the organic extract (~30-50 μl) are applied to a 5 x 20 cm analytical TLC plate, and separation is carried out using the same conditions that will be employed for the preparative TLC steps.
- If a pure standard of the particular N-coronafacoyl phytotoxin of interest is not available, then the N-coronafacoyl phytotoxin coronatine (COR) can be used instead. This phytotoxin is available for purchase from Sigma-Aldrich Canada (catalog number: C8115).
- For best results, make sure that the spotted extract has air dried before new extract is applied to the same area.
- The position of the N-coronafacoyl phytotoxin of interest on the reference analytical TLC plate will not correspond exactly to the phytotoxin position on the preparative TLC plate (as demonstrated in Figures 4 and 5). However, by comparing the relative position of the standard and extract bands on the reference plate, the location of the phytotoxin on the preparative TLC plate can be estimated. As the N-coronafacoyl phytotoxin of interest will not appear as a distinct band or fragment on the preparative TLC plate but as a dark smear, it is recommended that an area of at least 1 cm past the edge of this smear is marked.
- The sample should not be concentrated to a volume lower than this as otherwise the metabolites may precipitate out of solution.
- The HPLC running conditions described are suitable for separation of the CFA-L-Ile coronafacoyl phytotoxin from other metabolites in the extract. However, if minor N-coronafacoyl phytotoxins containing other isoleucine isomers (e.g., D-isoleucine) are produced, they will not be separated from the CFA-L-Ile metabolite and will be co-purified. Also, if other N-coronafacoyl phytotoxins or biosynthetic intermediates are being targeted for purification, the HPLC mobile phase running conditions may need to be adjusted accordingly in order to get good separation of those metabolites.
- Using the HPLC column and running conditions described, the CFA-L-Ile coronafacoyl phytotoxin has a retention time of 18-19 min (Figure 6).
- The purified metabolite can also be re-dissolved in MeOH + 0.1% formic acid to a final concentration of 1 mg/ml, and then stored at -20 °C.
- To confirm the purity of the metabolite, the sample can be analyzed using liquid chromatography-mass spectrometry (LC-MS) as described elsewhere (Bown et al., 2016).
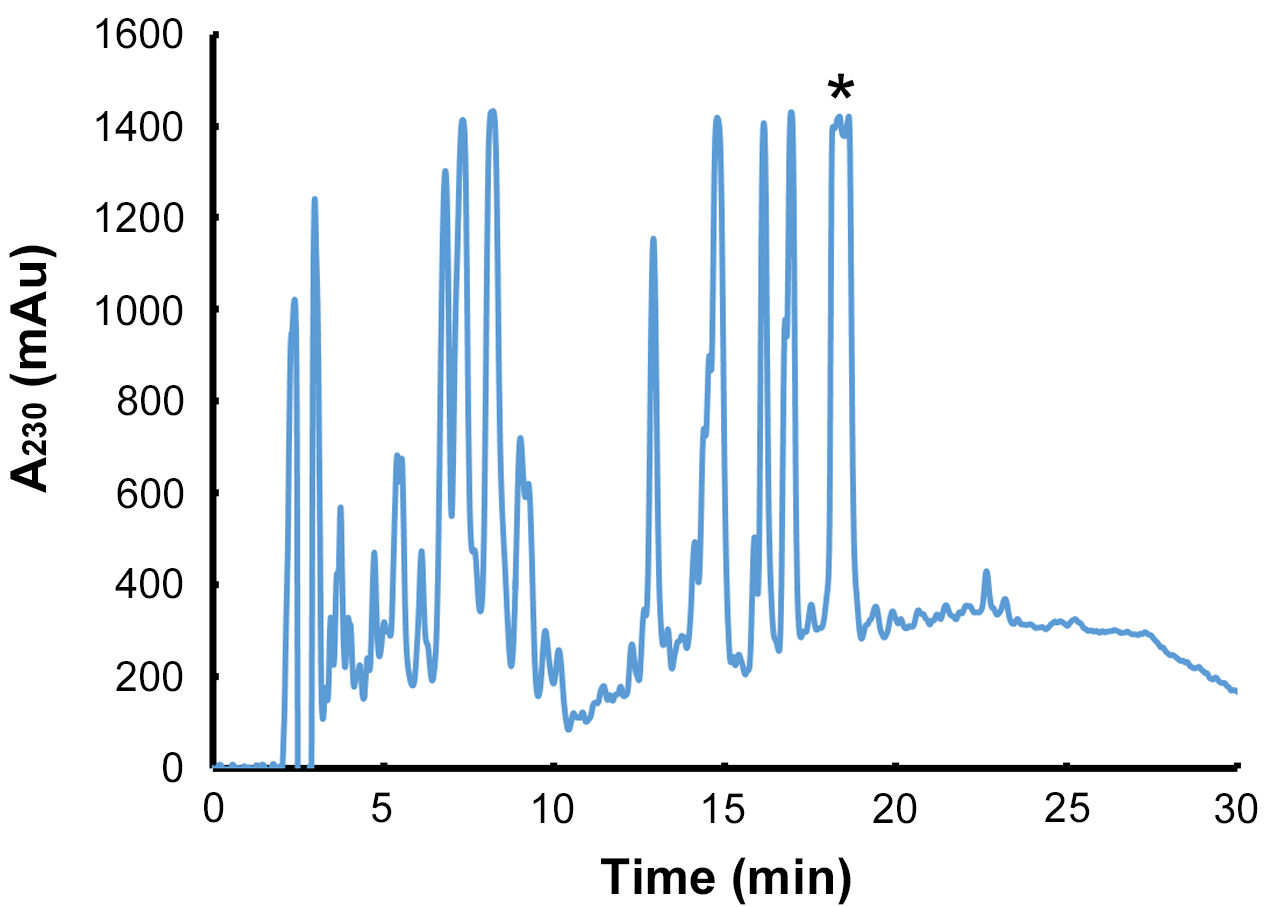
Figure 6. Purification of CFA-L-Ile by semi-preparative RP - HPLC. Following purification by preparative TLC, the CFA-L-Ile - containing extract is loaded onto a C18 semi-preparative HPLC column, and the metabolite is monitored by measuring the absorbance at 230 nm. The peak corresponding to CFA-L-Ile is indicated by the asterisks, and it is this peak that is targeted for collection by the HPLC fraction collector. Note that due to the high amount of CFA-L-Ile in the extract, the absorbance reading exceeds the maximum level measured by the detector, and thus the peak appears cut off in the chromatogram. Also, as N-coronafacoyl phytotoxins containing different isoleucine isomers can sometimes be produced by S. scabies in SFMB, the collected peak may contain a mixture of the different isomers since they will not separate out during the HPLC purification (Bown et al., 2016).
Recipes
- Soy flour mannitol broth (SFMB) medium (Kieser et al., 2000)
20 g/L soy flour
20 g/L D-mannitol
Dissolve mannitol in water
Add soy flour
Sterilize by autoclaving - Preparative TLC mobile phase (Fyans et al., 2015)
195 ml ethyl acetate
4 ml 2-propanol
0.5 ml acetic acid
0.5 ml water
Acknowledgments
This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC), the Canada Foundation for Innovation (CFI), and the Newfoundland and Labrador Research & Development Corporation (RDC) to D.R.D.B. L.B. was supported by a fellowship from the Memorial University School of Graduate Studies. This protocol is based on work originally reported in Bown et al. (2016) and Fyans et al. (2015).
References
- Bignell, D. R., Huguet-Tapia, J. C., Joshi, M. V., Pettis, G. S. and Loria, R. (2010). What does it take to be a plant pathogen: genomic insights from Streptomyces species. Antonie van Leeuwenhoek 98(2): 179-194.
- Bown, L., Altowairish, M. S., Fyans, J. K. and Bignell, D. R. (2016). Production of the Streptomyces scabies coronafacoyl phytotoxins involves a novel biosynthetic pathway with an F420 -dependent oxidoreductase and a short-chain dehydrogenase/reductase. Mol Microbiol 101(1): 122-135.
- Dees, M. W. and Wanner, L. A. (2012). In search of better management of potato common scab. Potato Research 55(3): 249-268.
- Fyans, J. K., Altowairish, M. S., Li, Y. and Bignell, D. R. (2015). Characterization of the coronatine-like phytotoxins produced by the common scab pathogen Streptomyces scabies. Mol Plant-Microbe Interact 28(4): 443-454.
- Mitchell, R. E. and Frey, E. J. (1986). Production of N-coronafacoyl-L-amino acid analogues of coronatine by Pseudomonas syringae pv. atropurpurea in liquid cultures supplemented with L-amino acids. Microbiology 132(6): 1503-1507.
- Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F. and Hopwood, D. A. (2000). Practical Streptomyces genetics. The John Innes Foundation.
- Palmer, D. A. and Bender, C. L. (1993). Effects of environmental and nutritional factors on production of the polyketide phytotoxin coronatine by Pseudomonas syringae pv. glycinea. Appl Environ Microbiol 59(5): 1619-1626.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Bown, L. and Bignell, D. R. D. (2017). Purification of N-coronafacoyl Phytotoxins from Streptomyces scabies. Bio-protocol 7(7): e2214. DOI: 10.21769/BioProtoc.2214.
Category
Microbiology > Microbial biochemistry > Other compound
Biochemistry > Other compound > Phytotoxin
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











