- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
3D Stroma Invasion Assay
Published: Vol 7, Iss 6, Mar 20, 2017 DOI: 10.21769/BioProtoc.2195 Views: 9337
Reviewed by: HongLok LungAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Determination of Ligand-Target Interaction in vitro by Cellular Thermal Shift Assay and Isothermal Dose-response Fingerprint Assay
Danyu Du [...] Jing Xiong
Aug 5, 2024 4953 Views

In Vitro Assay to Examine Osteoclast Resorptive Activity Under Estrogen Withdrawal
Cara Fiorino [...] Rene E. Harrison
Jan 5, 2025 1662 Views

Purification of Native Acetyl CoA Carboxylase From Mammalian Cells
Yaxue Sun [...] Hongtao Zhu
Feb 20, 2025 2193 Views
Abstract
We have developed a 3D co-culture system composed of fibroblasts and colorectal cancer cells that enables us to study the desmoplastic reaction. This method also enables us to study the influence of the desmoplastic reaction on the migration of colorectal cancer cells through the surrounding stroma. This protocol has been previously published (Coulson-Thomas et al., 2011) and is described here in more detail.
Keywords: 3D cultureBackground
The progression of cancer relies on intricate cross-talk between the cancer cells and surrounding cells, such as fibroblasts, inflammatory cells and endothelial cells, which form the cancer microenvironment. Fibroblasts are the major extracellular matrix producing cells and are responsible for the structural formation of tissues. Fibroblasts surrounding tumors are ‘activated’ by cancer cells into tumor-associated fibroblasts (TAFs) and play key roles in tumorigenesis and metastasis. In some cancers, TAFs up-regulate extracellular matrix expression producing an unorganized matrix, consisting mainly of collagen fibers and proteoglycans, which affects cancer cell proliferation, migration and spread. This is called the desmoplastic reaction, and during cancer cell growth different tumors may exhibit various grades of desmoplasia.
Materials and Reagents
- NuncTM Lab-TekTM II Chamber SlideTM System with 2 wells (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 154461 )
- NuncTM cell culture dishes (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 172931 )
- Cotton swabs
- Human colorectal fibroblasts CCD-112CoN (ATCC, catalog number: CRL-1541 )
- Caco-2 and HCT 166 cancer cell lines isolated from primary colorectal tumors (ATCC, catalog numbers: HTB-37 and CCL-247 )
- pEGFP-N1 (TaKaRa Bio, Clontech)
- DMEM culture medium (Thermo Fisher Scientific, GibcoTM)
- RPMI culture medium (Thermo Fisher Scientific, GibcoTM)
- Fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM)
- L-glutamine (Thermo Fisher Scientific, GibcoTM, catalog number: 25-030-081 )
- Penicillin/streptomycin (Thermo Fisher Scientific, InvitrogenTM)
- L-ascorbic acid (Sigma-Aldrich, catalog number: A4403 )
- Collagen I
- Anti-fibronectin (BD transduction laboratories)
- FuGENE® HD transfection reagent (Promega, catalog number: E2311 )
- Trypsin/EDTA (Thermo Fisher Scientific, GibcoTM)
- Paraformaldehyde, aqueous solution - 16% (Electron Microscopy Sciences, catalog number: 15700 )
- Complete media for Caco-2 and HCT 166 cells (see Recipes)
- Complete media for fibroblasts (see Recipes)
- Media for maintaining 3D cultures (see Recipes)
Equipment
- Ultra-fine forceps with a straight tip (Fine Science tools, catalog number: 11399-80 )
- CO2 cell culture incubator (Thermo Fisher Scientific, Thermo ScientificTM, model: HeracellTM 150i , catalog number: 51026280)
- Table top centrifuge (Eppendorf, model: 5702 RH )
- Vi-CELL XR cell counter (Beckman Coulter)
- Biological safety cabinets (Thermo Fisher Scientific, Thermo ScientificTM, model: Safe 2020 Class II , catalog number: 51026639)
- Scanning confocal inverted microscope (Zeiss, model: LSM 510 )
- Time-lapse confocal microscope (Zeiss, model: LSM 710 )
Software
- Java ImageJ and the Zen Imaging software from Zeiss
- Excel (Microsoft)
- GraphPad Prism (GraphPad Software)
Procedure
- Preparation of a 3D fibroblast-produced matrix
- Seed fibroblasts at a density of 3 x 106 cells per well of a 2 well-chamber slide to form the control 3D matrix and a mixture of fibroblasts and tumor cells at a density of 3 x 106 fibroblasts and 0.5 x 106 tumor cells per well to form the desmoplastic 3D matrix in DMEM and RPMI (1:1) containing 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin.
- Incubate the cells at 37 °C in a 5% CO2 humidified environment until confluent.
- Once a confluent monolayer is formed, treat the cultures with 3 ml of 25 µg/ml ascorbic acid every other day for an additional 10 days to induce the production of a 3D matrix, maintaining the cells at 37 °C in a 5% CO2 humidified environment. The medium should be changed every other day with fresh medium containing 25 µg/ml ascorbic acid. The cells may be treated for longer than 10 days if a thicker and denser matrix is desired. Extracellular matrix components such as collagen I and fibronectin can be used to visualize the 3D matrix (Figure 1). All fibroblasts would respond to ascorbic acid treatment; however, the thickness, density and organization may vary among different types of fibroblasts.
Note: Fresh ascorbic acid solutions should be prepared every other day since ascorbic acid in solution decays over time.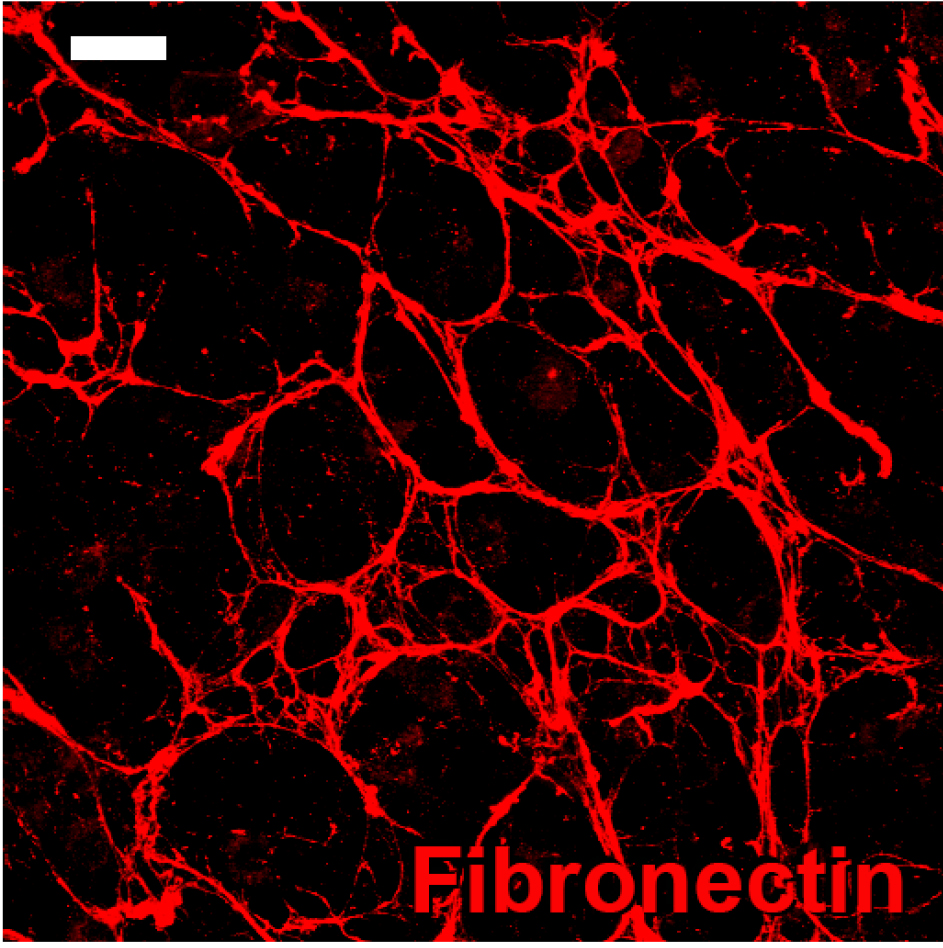
Figure 1. Fibronectin immunostaining with mouse anti-fibronectin evidencing 3D matrix formation in a 3D stromagenic model. Scale bar = 20 μm.
- Seed fibroblasts at a density of 3 x 106 cells per well of a 2 well-chamber slide to form the control 3D matrix and a mixture of fibroblasts and tumor cells at a density of 3 x 106 fibroblasts and 0.5 x 106 tumor cells per well to form the desmoplastic 3D matrix in DMEM and RPMI (1:1) containing 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin.
- Cell invasion assay
- Transfect colorectal tumor cells with the GFP plasmid using FuGENE® HD transfection reagent (500 µg of plasmid in a total volume of 2 ml) and incubate the cells at 37 °C in a 5% CO2 humidified environment for 48 h, according to the manufacturer’s instructions.
Note: It is highly recommended for the first experiment to test the best confluence at which the cells should be transfected. This would entail seeding cells at increasing densities, for example at 30%, 50%, 75% and 90% confluence, in order to test which confluence provides the highest transfection rate. We established that for us 75% confluence provided the highest transfection rate. - Remove the GFP transfected cells from the culture dish using trypsin/EDTA and seed 1 x 106 tumor cells onto the 3D stromagenic systems prepared in item A (both experimental and control systems – see Notes below).
- Allow the GFP positive cells to migrate for 1 h, maintaining the cells at 37 °C in a 5% CO2 humidified environment after which the invading cells can be imaged using time-lapse confocal microscopy.
- Carry out the time-lapse analysis using a scanning confocal inverted microscope. Time-lapse images can be captured every minute for a total of 15 to 30 min to enable the investigator to see the movement of the tumor cells invading the 3D matrix.
- Z-stacks should be obtained throughout the thickness of the 3D matrix at the end of the 2 h period to verify the depth to which the cancer cells invaded, as previously described (Coulson-Thomas et al., 2011; de Paula et al., 2012). Z-stack projections using the z-axis as reference can be made using any imaging software, such as Image Processing and Analysis with Java ImageJ and the Zen Imaging software from Zeiss.
- Transfect colorectal tumor cells with the GFP plasmid using FuGENE® HD transfection reagent (500 µg of plasmid in a total volume of 2 ml) and incubate the cells at 37 °C in a 5% CO2 humidified environment for 48 h, according to the manufacturer’s instructions.
Data analysis
The time-lapse images can be assembled into a video using the LSM or ZEN software. A z-stack of the images may also be projected using the LSM or ZEN software in order to estimate the depth of the invading cell. We carried out our experiments three times in triplicate, and our statistical analyses were calculated using the t-test in Excel (Microsoft) and GraphPad Prism (GraphPad Software).
Notes
- Experimental groups consist of seeding GFP positive colorectal tumor cells onto the desmoplastic 3D stromagenic system composed of fibroblasts and colorectal cancer cells, while control groups consist of seeding GFP positive colorectal tumor cells onto the 3D stromagenic system composed of solely fibroblasts.
- This protocol was developed to study the invasion of colorectal tumor cells, but any other cancer cell type could be tested.
- The two-hour migration period should be tested and adapted if necessary when using different cancer cell lines.
- The same procedure can be used to induce the production of a 3D fibroblast-produced matrix in culture plate inserts with a pore size of 8 µm (30 mm, Millicell®-PCF, Millipore Corp). In this case, the GFP positive colorectal tumor cells are seeded on the 3D stromagenic matrix in the culture plate insert in serum free medium and allowed to migrate for 8 h to the lower chamber containing serum supplemented with 10% FBS. Once fixed using 10% paraformaldehyde, cells are removed from the upper compartment using a cotton swab and the GFP positive cells that migrated are analyzed.
Recipes
- Complete media for Caco-2 and HCT 166 cells
RPMI medium supplemented with 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin - Complete media for fibroblasts
DMEM medium supplemented with 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin - Media for maintaining 3D cultures
A mixture of RPMI and DMEM media (1:1) supplemented with 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin and 25 μg/ml ascorbic acid
Acknowledgments
This protocol is from Coulson-Thomas et al., 2011. This work was supported by CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) and FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo).
References
- Coulson-Thomas, V. J., Coulson-Thomas, Y. M., Gesteira, T. F., de Paula, C. A., Mader, A. M., Waisberg, J., Pinhal, M. A., Friedl, A., Toma, L. and Nader, H. B. (2011). Colorectal cancer desmoplastic reaction up-regulates collagen synthesis and restricts cancer cell invasion. Cell Tissue Res 346(2): 223-236.
- de Paula, C. A., Coulson-Thomas, V. J., Ferreira, J. G., Maza, P. K., Suzuki, E., Nakahata, A. M., Nader, H. B., Sampaio, M. U. and Oliva, M. L. (2012). Enterolobium contortisiliquum trypsin inhibitor (EcTI), a plant proteinase inhibitor, decreases in vitro cell adhesion and invasion by inhibition of Src protein-focal adhesion kinase (FAK) signaling pathways. J Biol Chem 287(1): 170-182.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Coulson-Thomas, Y. M. and Coulson-Thomas, V. J. (2017). 3D Stroma Invasion Assay. Bio-protocol 7(6): e2195. DOI: 10.21769/BioProtoc.2195.
Category
Cancer Biology > Invasion & metastasis > Tumor microenvironment
Cancer Biology > Cancer biochemistry > Protein
Cell Biology > Cell-based analysis > Extracellular microenvironment
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










