- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Isolation of Outer Membrane Vesicles from Phytopathogenic Xanthomonas campestris pv. campestris
Published: Vol 7, Iss 5, Mar 5, 2017 DOI: 10.21769/BioProtoc.2160 Views: 12841
Reviewed by: Zhaohui LiuEmilie ViennoisMahmoud Kamal Ahmadi

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In Silico Prediction and In Vitro Validation of Bacterial Interactions in the Plant Rhizosphere Using a Synthetic Bacterial Community
Arijit Mukherjee [...] Sanjay Swarup
Nov 5, 2025 1675 Views

Preparation and Negative Staining for Visualization of Cyanoglobule Lipid Droplets Using Transmission Electron Microscopy
Febri A. Susanto [...] Peter K. Lundquist
Dec 5, 2025 1316 Views

On-Column Dual-Gradient Refolding for Efficient Recovery of Insoluble Affinity-Tagged Recombinant Proteins
Anna Vlaskina [...] Maxim Patrushev
Feb 5, 2026 92 Views
Abstract
Gram-negative bacteria naturally release outer membrane vesicles (OMVs) to the surrounding environment. OMVs contribute to multiple processes, such as cell-cell communication, delivery of enzymes and toxins, resistance to environmental stresses and pathogenesis. Little is known about OMVs produced by plant-pathogenic bacteria, and their interactions with host plants. The protocol described below discusses the isolation process of OMVs from Xanthomonas campestris pv. campestris strain 33913, a bacterial pathogen of Crucifiers. Nevertheless, this protocol can be used and/or adapted for isolation of OMVs from other phytopathogenic bacteria to promote the study of OMVs in the context of plant-microbe interactions.
Keywords: OMVsBackground
Extracellular vesicle (EV) release is a process shared by many organisms from all domains of life. In Gram-negative bacteria, most EVs are a result of the outer membrane blebbing and eventually pinching off the bacterial cell wall, and are hence referred to as outer membrane vesicles (OMVs). The study of OMVs focuses on OMV biogenesis, cargo, functions and interactions with host organisms. To date, most of the study on OMVs focused on bacterial pathogens of humans and environmental bacteria, however very little research has been done on OMVs from phytopathogenic bacteria. The protocol described here was adapted from the protocol described by Chutkan et al. (2013) with slight modification, and presented here with the phytopathogen X. campestris pv. campestris. To our understanding this is the first, fully detailed, protocol for isolation of OMVs from phytopathogenic bacteria and we hope it could serve as a guiding protocol for other research groups interested in this topic.
Materials and Reagents
- Sterile single-use inoculating loop
- 200 μl pipette tips (such as: Corning, Axygen®, catalog number: # T-200-C )
- Parafilm (Bemis, catalog number: PM996 )
- Syringe-mounted 0.45-µM filter (EMD Millipore, catalog number: SLHV033RS )
- Polyethersulfone (PES) filter, 0.45-µM, Ø75-mM membrane diameter, with an attached 500-ml bottle. Operated by a vacuum pump (e.g., Guangzhou Jet Bio-Filtration, catalog number: FPE404500 )
- 1 ml and 10 ml sterile syringes (without needle)
1 ml syringes (such as: KDL Medical Product, catalog number [as written on the package]: 7290010993604)
10 ml syringes (such as: Medisposables, catalog number [as written on the package]: 9304-10/16933506093041 ) - Sterile 15-ml Falcon tube (such as: SPL Life Sciences, catalog number: 50015 )
- 12 ml polypropylene/polyallomer ultracentrifuge tubes (such as: Beckman Coulter, catalog number: 331372 ) – make sure that they are fit for the ultracentrifuge’s speed
- Syringe-mounted 0.22-µM filter (Axiva Sichem Biotech, catalog number: SFPV13 R )
- Sterile 1.5-ml tube (1.7-ml clear tube) (Corning, Axygen®, catalog number: # MCT-175-C )
- 1 ml pipette tips (such as: Corning, Axygen®, catalog number: # T-1000-C )
- Polystyrene (PS) aseptic Petri dishes, 90 x 15-mm (Miniplast Ein-Shemer, catalog number: 820-090-01-017 )
- Xanthomonas strain in glycerol stock stored at -80 °C – for the preparation of this protocol we used X. campestris pv. campestris (Xcc) 33913, which was obtained from American Type Culture Collection (ATCC)
- Difco nutrient agar (BD, catalog number: 21300 )
Note: It contains 3 g/L of beef extract, 5 g/L of peptone, and 15 g/L of agar, with a final pH of 6.8 ± 0.2 (as written on the powder’s container). - Cephalexin hydrate (Sigma-Aldrich, catalog number: C4895 ) – stock solution at concentration of 10 mg/ml
- Sterile dH2O
- 10x phosphate buffered saline (PBS) buffer (optional) (such as: Sigma-Aldrich, catalog number: P5493 )
- OptiPrep density gradient medium (Sigma-Aldrich, catalog number: D1556 )
- ‘Coomassie Plus – The Better Bradford Assay Kit’ (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 23236 ) (optional)
- Lipid dye FM4-64FX (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: F34653 ) (optional)
- Materials and reagents required for sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), as indicated in the following protocol (optional): http://www.bio-protocol.org/e80 (‘[Bio101] Laemmli-SDS-PAGE’)
- Yeast extract powder (EMD Millipore, catalog number: 61931105001730 )
- Bacto tryptone (BD, Bacto, catalog number: 211705 )
- Sodium chloride (NaCl) (EMD Millipore, catalog number: 7647-14-5 )
- Sucrose crystallized (Duchefa Biochemie, catalog number: S0809 )
- Magnesium sulfate heptahydrate (MgSO4·7H2O) (EMD Millipore, catalog number: 105886 )
- NaOH or HCl (in order to adjust pH)
- Peptone (EMD Millipore, catalog number: 61930705001730 )
- L(+)-glutamic acid (Acros Organics, catalog number: 6106-04-3 )
- HEPES (Sigma-Aldrich, catalog number: H3375 )
- YEB medium (see Recipes)
- PSB medium (see Recipes)
- OptiPrep diluent buffer (see Recipes)
Note: (optional) – not necessary for the vesicles’ isolation process.
Equipment
- Biological hood
- Temperature controlled incubator shaker (28 °C), with a capability to reach 200 RPM (such as: Thermo Fisher Scientific, Thermo ScientificTM, model: MaxQTM 6000 )
- Two 2-L Erlenmeyer flasks
- Cuvette
- Polypropylene Copolymer (PPCO) 250-ml centrifuge bottles (such as: Thermo Fisher Scientific, Thermo ScientificTM, model: NalgeneTM PPCO Centrifuge Bottle ) – make sure that they are fit for the centrifuge’s speed
- High-speed centrifuge (e.g., Beckman Coulter, model: Avanti J-E , catalog number: 369003; or Thermo Fisher Scientific, model: SorvallTM RC-6 , catalog number: 74804*) and 6 x 250-ml rotor (such as: Beckman Coulter, model: JLA-16.250 , catalog number: 363930, dual-locking lid; or Thermo Fisher Scientific, SorvallTM, model: SLA-1500 , catalog number: 46615* [old model]) at a speed of approx. 32,000 x g
- Ultracentrifuge (e.g., Thermo Fisher Scientific, SorvallTM, model: Discovery 90SE , catalog number: 40130867*, Kendro Laboratory Products) and a 6 x 12-ml swinging bucket ultracentrifuge rotor such as: Thermo Fisher Scientific, SorvallTM, model: TH-641 , catalog number: 54295* (old model, Kendro Laboratory Products), or Thermo Fisher Scientific, Thermo ScientificTM, model: TH-641 , catalog number: 54295 (new model,) at a speed of approx. 175,000 x g
- Autoclave
- NanoSight device (Malvern) (optional)
- Transmission electron microscopy (optional)
- Equipment required for SDS-PAGE as indicated in the following protocol (optional): http://www.bio-protocol.org/e80 (‘[Bio101] Laemmli-SDS-PAGE’)
Notes
- *Catalog number of an old model.
- (optional) – not necessary for the vesicles’ isolation process.
- Important note: the centrifuge speeds mentioned above, were calculated with Science Gateway's centrifuge rotor speed calculator (http://www.sciencegateway.org/tools/rotor.htm) – if the centrifuge/ultracentrifuge did not give such option.
Procedure
- Preparations prior to OMV isolation
- Preparation of 1 L of YEB (for starters, see Recipes).
- Preparation of 1 L of PSB (the main medium, see Recipes).
- Plating the bacteria
Plate Xcc 33913 (from glycerol stock) on Difco nutrient agar plates, by using a sterile single-use inoculating loop.
Note: It is recommended to plate the bacteria 2-5 days before making the starters with Xcc. - Making Xcc starters (2 sterile tubes with 3 ml of sterile YEB in each)
- To every tube, add 3 µl of cephalexin hydrate 10 mg/ml (final concentration will be 10 µg/ml of medium).
- Using a sterile 200-µl pipette tip, scrub 3-5 Xcc colonies from the plate, and add them to the medium in the tube. Must be done in a biological hood.
- Put the tubes in 28 °C shaker for overnight (the speed is 180-200 rpm).
- Inoculation of the main medium (PSB)
- To every 500 ml of PSB medium in each 2-L Erlenmeyer flask, add 500 µl of cephalexin hydrate 10 mg/ml (final concentration will be 10 µg/ml of medium).
- Then, from one of the Erlenmeyer flasks, transfer 1 ml of medium (+ cephalexin) into a clean, new cuvette (will serve as a blank for the spectrophotometer). Cover the cuvette with 2 layers of Parafilm, and keep the cuvette at 4 °C.
- Then, to each Erlenmeyer flask, add 300 µl from the same starter (has to be very cloudy).
- Put the Erlenmeyer flasks into 28 °C shaker, at the speed of 180-200 rpm, for overnight (Video 1).
- To every 500 ml of PSB medium in each 2-L Erlenmeyer flask, add 500 µl of cephalexin hydrate 10 mg/ml (final concentration will be 10 µg/ml of medium).
- Must be done in a biological hood.
- The ‘cloudiness’ of the starter might affect the time needed for the inoculated PSB media, to reach the desired OD600 nm.
- Temperature above 30 °C might inhibit bacterial growth in the Erlenmeyer flasks.Video 1. Culture incubation
- On the next day – object density measurement
- The cuvette stored at 4 °C (with the PSB medium and cephalexin hydrate) serves as a blank. This cuvette should be kept at 4 °C between measurements.
- At the ‘normal range’ described in Table 1, the concentration of Xcc 33913 is approximately 108 cfu/ml, and the cells are at around the mid log phase of the growth curve. Just before cell harvest, a small sample of the culture is taken, serially diluted and plated to determine cell number and to assess the amount of purified vesicles per cell.
Table 1. The object density ranges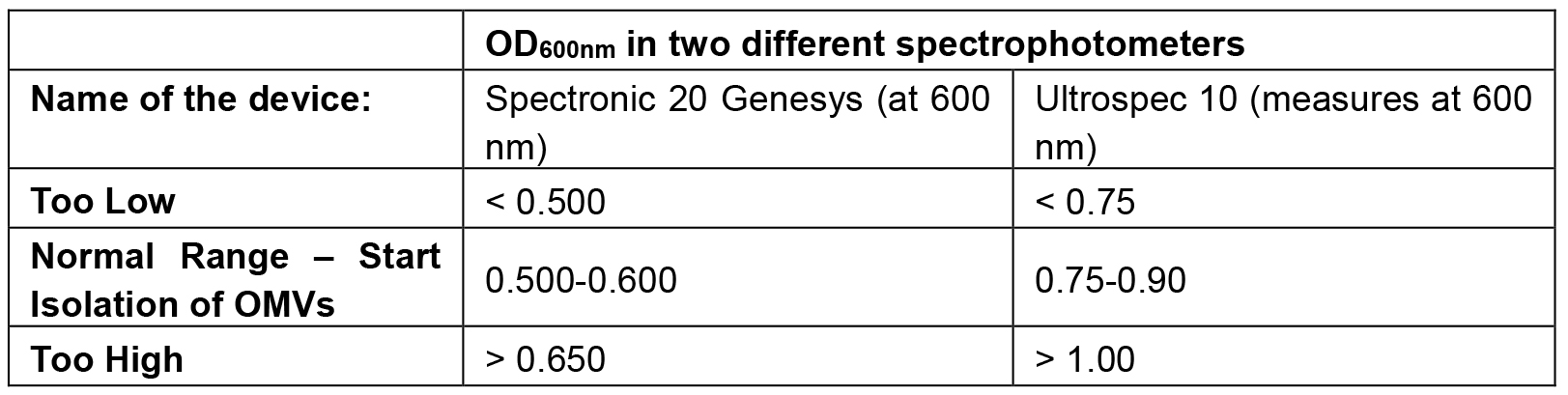
- The cuvette stored at 4 °C (with the PSB medium and cephalexin hydrate) serves as a blank. This cuvette should be kept at 4 °C between measurements.
- Must be done in a biological hood. Since different spectrophotometers may give different reads for the same sample, it is recommended that first a proper bacterial growth curve experiment will be conducted using OD measurements and cell counts on plates, to accurately define the different growth phases and their respective ODs with the instrument used.
- About the required OD600 nm: Ideally, cells should be harvested at mid-log phase. Harvesting at a later time point might result in cell death and cell debris, and/or release of other substances that could contaminate the preparation and/or clog the filtering membrane. The typical incubation time, that is required in order to get to the desired OD600 nm, is 17-21 h.
- Preparation of 1 L of YEB (for starters, see Recipes).
- The isolation process
- Removal of bacteria
- Pour the cultures into 250-ml bottles, and centrifuge them at approx. 13,200 x g for 22 min, at the temperature of 4 °C to pellet bacterial cells. Rotors that can be used: GSA (Sorvall), JLA-16.250 (Beckman-Coulter) (Video 2). Video 2. Removal of bacteria
- Then, filter the supernatant through a 0.45-µM PES filter with Ø75-mM membrane diameter (operated by a vacuum pump).
Optional: It is possible to filter the entire 1 L of the culture through such filter; thus, you can connect it to a 1-L bottle (if the filter fits). The filtered supernatant can be stored at 4 °C up to 1 day.
- Pour the cultures into 250-ml bottles, and centrifuge them at approx. 13,200 x g for 22 min, at the temperature of 4 °C to pellet bacterial cells. Rotors that can be used: GSA (Sorvall), JLA-16.250 (Beckman-Coulter) (Video 2).
- Pelleting the vesicles
- Pour the filtered supernatant into 250-ml bottles, and centrifuge them at approx. 32,000 x g for 2 h, at 4 °C. The vesicles will be pelleted (Figure 1) in the outward direction from the center of the rotor. Rotors that can be used: SLA-1500 (Sorvall), JLA-16.250 (Beckman-Coulter) (Video 3).
- Discard the supernatant and resuspend the pelleted vesicles, using 5-8 ml of sterile dH2O, by pipetting.
Optional: Filter the resuspended vesicles through 0.45-µM (operated by a 10-ml syringe) into a sterile 15-ml Falcon tube, so you can keep them at 4 °C for a few days. Must be done in a biological hood.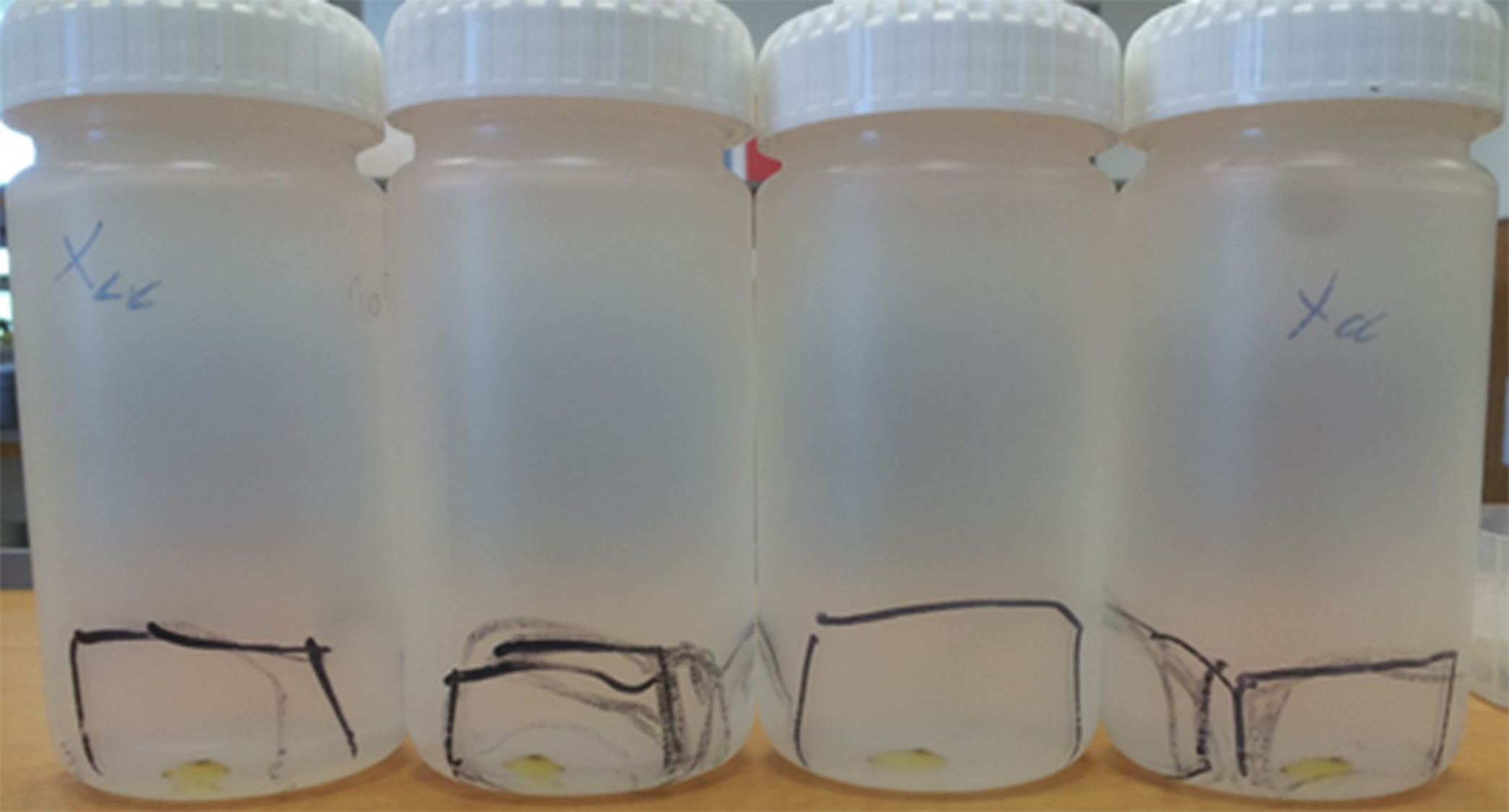
Figure 1. The pelleted OMVs with a translucent yellow color can be seen in the bottom of the tubes (within the marked area of each tube), after centrifugation at approx. 32,000 x g for 2 h
Video 3. Pelleting the vesicles
- Pour the filtered supernatant into 250-ml bottles, and centrifuge them at approx. 32,000 x g for 2 h, at 4 °C. The vesicles will be pelleted (Figure 1) in the outward direction from the center of the rotor. Rotors that can be used: SLA-1500 (Sorvall), JLA-16.250 (Beckman-Coulter) (Video 3).
- Ultracentrifugation
- Pour the resuspended vesicles, from the previous step, into two 12-ml ultracentrifuge tubes, and use sterile dH2O to fill the remaining volume of the tubes.
- Ultracentrifuge the resuspended vesicles, at approx. 175,000 x g, for 1 h and 35 min, at 4 °C. Rotor that can be used: Sorvall TH-641. Ultracentrifuge that can be used: Sorvall Discovery 90SE. The vesicles will be pelleted in the bottom of the tubes (Figure 2, Video 4).
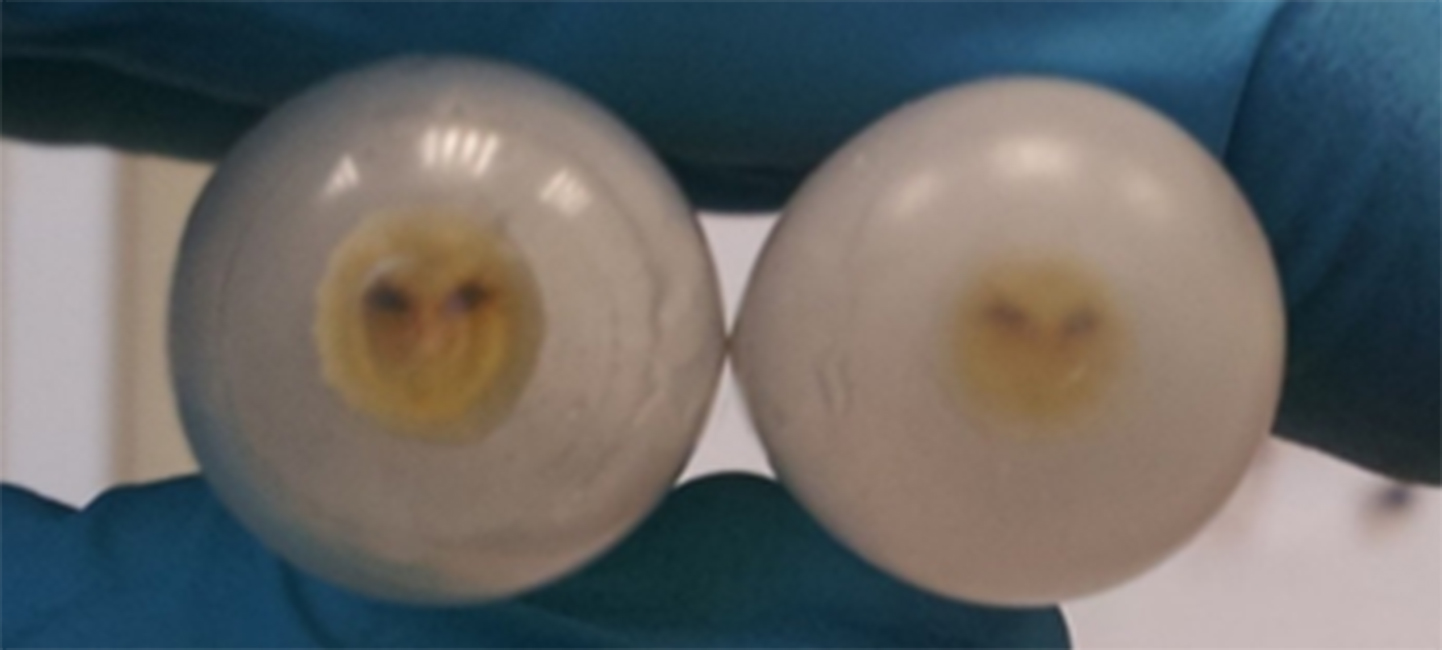
Figure 2. The pelleted OMVs seen in the bottom of the 12-ml tubes, after ultracentrifugation at approx. 175,000 x g for 1 h and 35 minVideo 4. Ultracentrifugation
- Pour the resuspended vesicles, from the previous step, into two 12-ml ultracentrifuge tubes, and use sterile dH2O to fill the remaining volume of the tubes.
- Final resuspension and storage
- Discard the supernatant and resuspend the pelleted vesicles with sterile dH2O or with PBS buffer by pipetting, using a volume that is 1/1,000 of original medium volume. For example: 1 ml of sterile dH2O for 1 L of PSB.
- Filter the resuspended vesicles through a 0.22-µM filter (operated by a syringe), into a sterile 1.5-ml tube. Must be done in a biological hood. The filtered OMV preparation can be stored at 4 °C as is or further purified by density gradient centrifugation.
- Density gradient centrifugation: In order to purify the OMV further, for analyses that require highly-purified samples (e.g., protein analysis in LC-MS/MS), a density gradient centrifugation step is required (Chutkan et al., 2013).
- For density gradient centrifugation, use OptiPrep density gradient medium 60% stock solution (Sigma-Aldrich), and make 45%, 40%, 35%, 30%, 25%, and 20% OptiPrep solutions (using OptiPrep diluent buffer, see Recipes).
- Carefully layer 1 ml of each diluted OptiPrep solution on the top of the other, starting from 45% OptiPrep solution at the bottom. On the top of the 20% OptiPrep solution, carefully layer the OMV prep and sterile dH2O.
Notes: - If you have the entire OMV sample layered in one tube, you must have another, balancing tube, with the same diluted OptiPrep layers, but a sterile dH2O layered on them (instead of OMV).
- It’s recommended to use equally-weighted tubes, even when empty, to avoid imbalance, and to keep equal volumes of the top layer (OMV or dH2O) between the tubes.
- Spin the samples at approx. 246,000 x g for 6 h, at 4 °C. After the ultracentrifugation, a layer/s of the vesicles will be visible in the tube, due to Xanthomonas yellow pigment Xanthomonadin (Figure 3; Video 5).
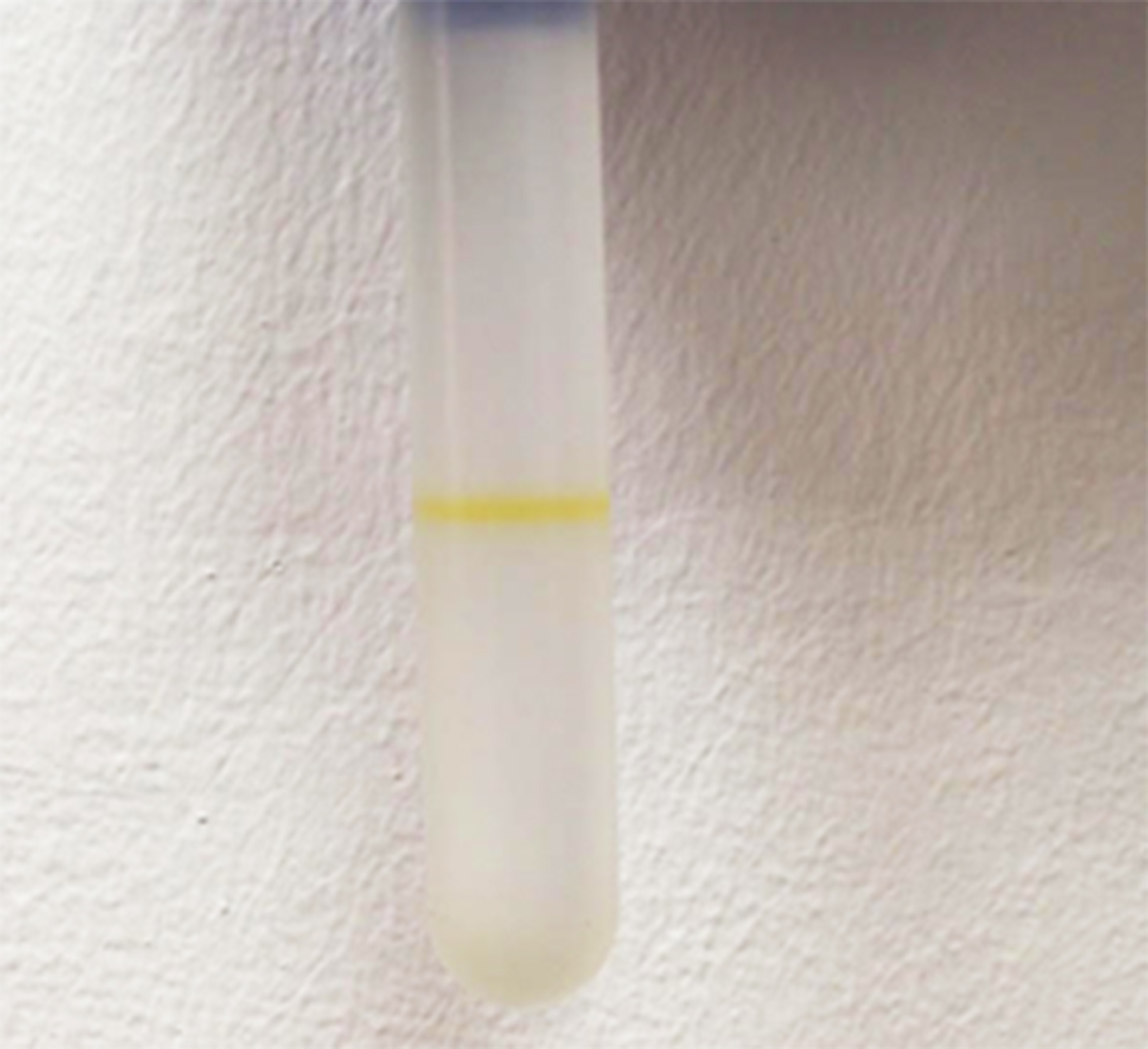
Figure 3. OMVs (the yellow line within the tube) are suspended within one of the fractions of the density gradient, after ultracentrifugation at approx. 246,000 x g for 6 hVideo 5. Purification by a density gradient - With a 1-ml pipette tip laid against the side of tube, slowly and carefully collect each 1 ml fraction from top to bottom. Each 1 ml collected should be transferred into a separate 12-ml ultracentrifuge tube. Change the pipette tip between the fractions.
- To each 12-ml ultracentrifuge tube containing a 1 ml fraction from the previous step (from the OptiPrep layers), add 11 ml of sterile dH2O to dilute the OptiPrep, and spin at approx. 170,000 x g for 2 h, at 4 °C (rotor that can be used: Sorvall TH-641; Ultracentrifuge: Sorvall Discovery 90SE).The vesicles will be pelleted
- Then, the pellet can be resuspended in 0.2-0.5 ml of sterile dH2O or PBS buffer and filtered (by a syringe-mounted 0.22-µM filter), or the pelleted OMV can be scrubbed from the bottom of the tubes by using two clean thin spatulas. This can be achieved by collecting (scrubbing) the pellet using one of the spatulas.
- Then, use the other spatula to remove the pellet from the first spatula and stick it, or smear it, on the sterile 1.5-ml tube’s wall. The 1.5-ml tube containing the pelleted OMVs can be then stored at -80 °C.
Note: Must be done in a biological hood.
Table 2. Isolation and purification process summary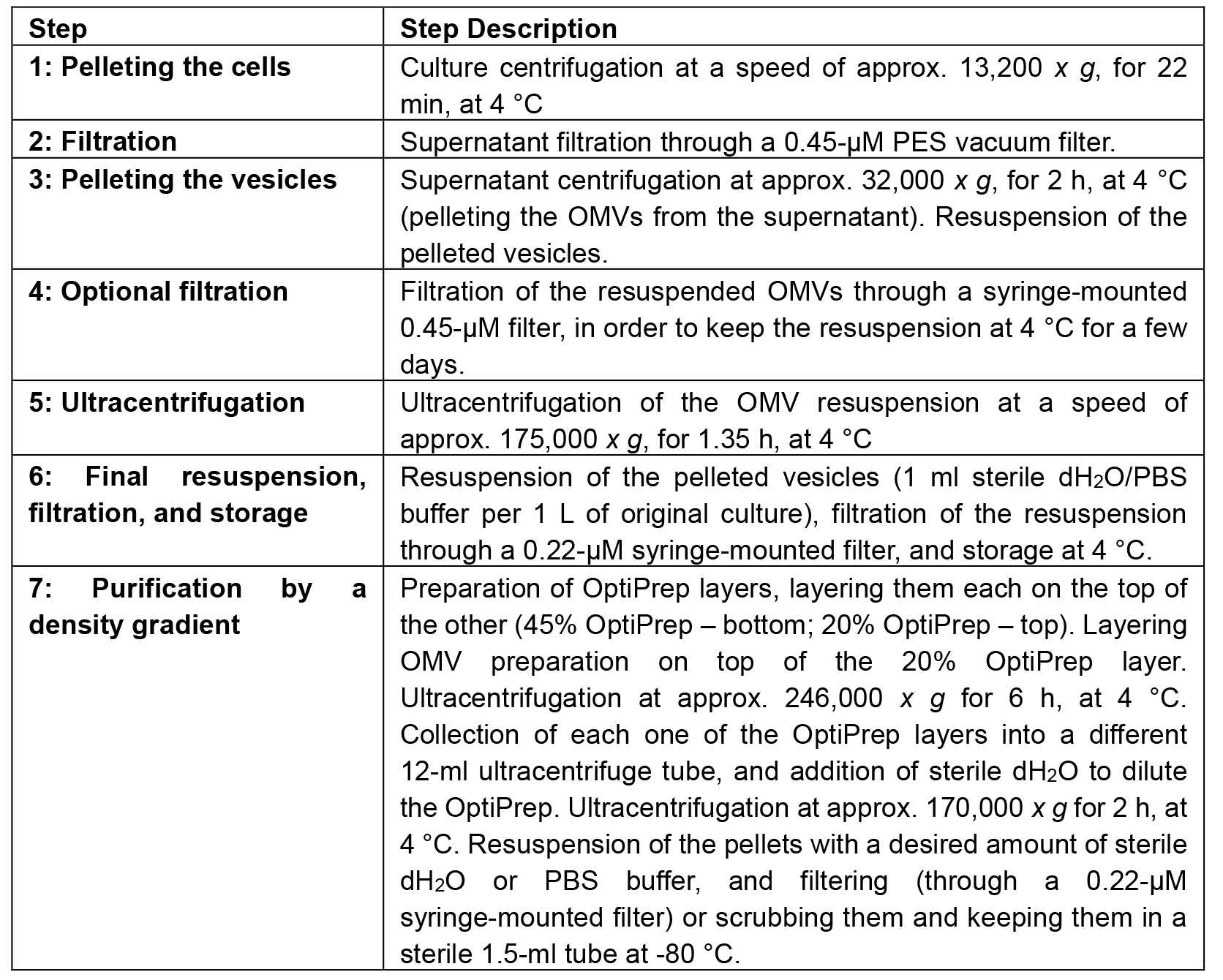
- Removal of bacteria
- OMV preparation quality and quantity checks
- Checking if the preparation is sterile
- Pipette 10 µl of OMV preparation on a plate with Difco nutrient agar, and leave open until the 10 µl drop dries out. Must be done in a biological hood.
- Then, place the plate in a 28 °C incubator for 2-5 days, and check if colonies are formed where the drop was placed.
- Pipette 10 µl of OMV preparation on a plate with Difco nutrient agar, and leave open until the 10 µl drop dries out. Must be done in a biological hood.
- Quantification of the OMVs
- ‘Coomassie Plus – The Better Bradford Assay Kit’, can be used to quantify the protein cargo of the vesicles.
- A NanoSight device, can be used to test the size distribution of the vesicles and their quantity.
- Another way to estimate the vesicles’ concentration, provided that the OMVs are from Xanthomonas, is by measuring the object density of Xanthomonadin. Xanthomonadin maximum absorbance is at 445 nm. When measuring OD445 nm, use sterile dH2O as blank (Munhoz et al., 2011; Goel et al., 2001).
Note: The OD445 nm of the OMV preparation differs between a filtered and a non-filtered preparation. - The lipid dye FM4-64FX can be used to estimate the concentration of the vesicles, based on lipid amount.
- Transmission electron microscopy (TEM)
TEM can be used to view OMV (Figure 4) and estimate size range and concentration as well as examine the purity of the preparation.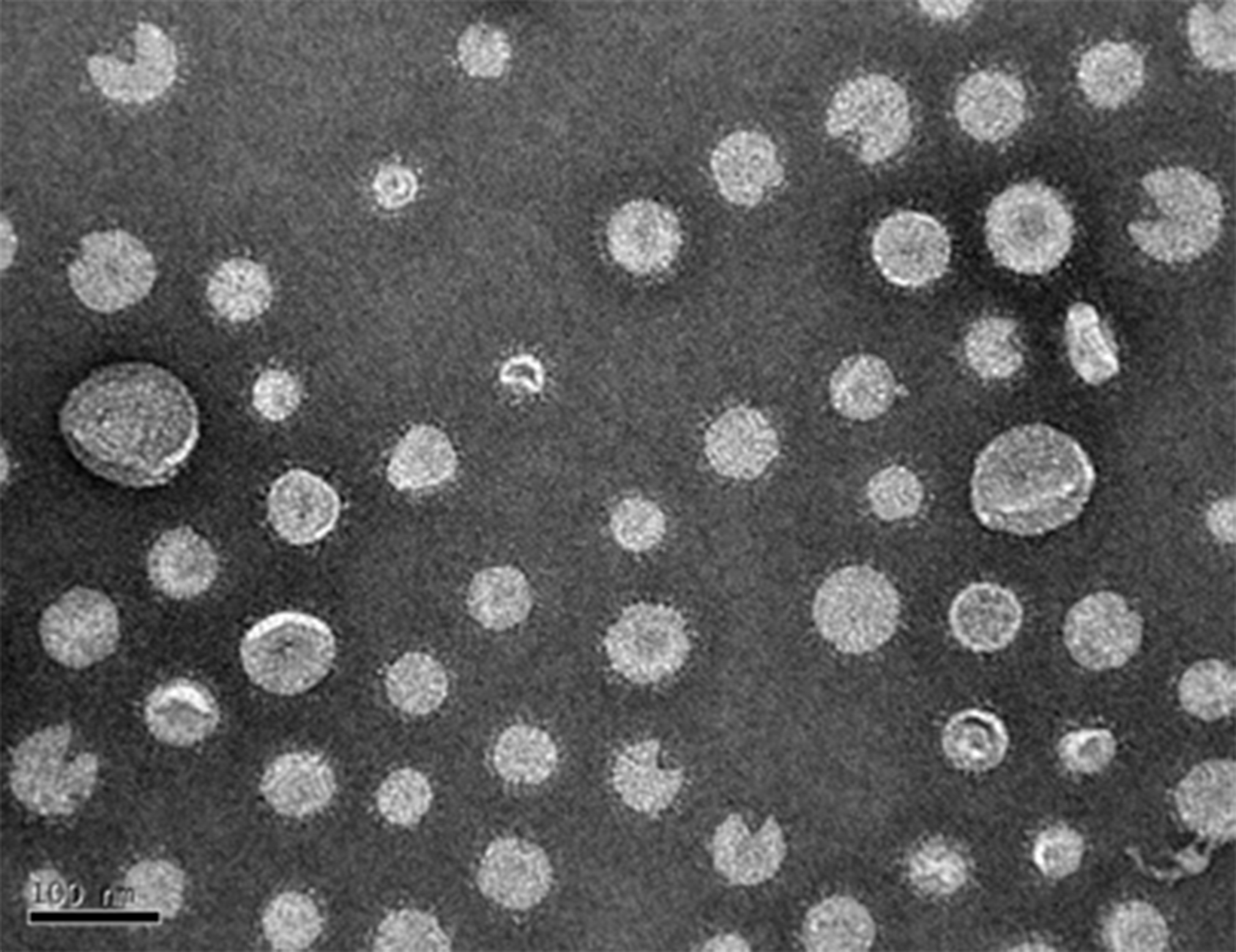
Figure 4. TEM image of an OMV preparation, diluted in sterile dH2O by a ratio of 1:20. OMV diameter may range from 20-250. A scale bar is at the bottom left. - SDS-PAGE
In order to get the profile of the proteinaceous cargo of OMV, run an OMV sample, which was denatured in a sample buffer containing sodium dodecyl sulfate (SDS) and dithiothreitol (DTT)/β-mercaptoethanol, on a protein gel. If you are searching for a specific protein associated with OMV, you can also run a Western blot or a dot blot assays. SDS-PAGE is also a way to compare the proteinaceous profiles of OMV isolated from different bacteria.
- Checking if the preparation is sterile
Data analysis
Perhaps one of the main challenges in purifying OMVs from bacterial culture is to prove its purity. Since OMVs are purified from culture supernatants the risk of contamination by surface, extracellular bacterial appendages, such as flagella, pili, fimbriae, must be taken into consideration. Carefully examining multiple TEM grid samples and determining whether such appendages had co-purified with the OMVs or not is highly recommended to assure purity. Another possible contaminant of OMV preparations are broken cell debris and large protein complexes. To minimize the chances of broken cell contamination cultures should be harvested during the logarithmic growth phase when cell death and breakage is less likely to occur. Additionally, we highly recommend that the density gradient centrifugation step will be carried out for any down-stream studies, especially for analytical studies such as proteomics, to avoid co-purification of large protein or protein complexes from the supernatant.
Recipes
- YEB (1 L, for starters)
- Add:
5 g of yeast extract powder
10 g of Bacto tryptone
5 g of NaCl
5 g of sucrose crystallized
5 g of MgSO4·7H2O
to 900 ml of dH2O - Adjust the pH to 7.3 using NaOH or HCl
Note: Do not heat while measuring pH. - Bring to 1 L by adding more dH2O, and autoclave at 121 °C for 20 min
- PSB (1 L, the main medium)
- Add:
10 g of peptone
10 g sucrose crystallized
1 g of L(+)-glutamic acid
to 900 ml of dH2O
A slight heating while stirring might assist in dissolving the components, yet it’s not compulsory. Turn off the heat, and transfer the beaker to another, colder, stirrer - Adjust the pH to 7.3 using NaOH or HCl
Note: Do not heat while checking pH. - Add more dH2O in order to reach 1 L
- Mix by passing the medium between the beaker and a measuring cylinder
- Divide the medium between two 2-L Erlenmeyer flasks (500 ml in each), and autoclave at 121 °C for 20 min
- OptiPrep diluent buffer (1 L)
- Add:
8.5 g of NaCl
2.38 g of HEPES
to approx. 750 ml of dH2O in a beaker - Adjust the pH to 7.4 with NaOH
Note: Do not heat while checking pH. - Bring to 1 L with dH2O and filter-sterilize with 0.45-μM filter (operated by a vacuum pump)
- Suggestion: keep the buffer at 4 °C
Acknowledgments
Work at O. Bahar lab was supported by the German-Israeli Foundation for Scientific Research and Development (GIF), grant No. I-2392-203.13/2015 and by the Israel Science Foundation, grant No. 2025/16. We would like also to thank our lab members for every help given, S. Burdman’s lab, M. Levy’s lab, and N. Sela, for collaboration in experiments, and also M. Mawassi’s lab, V. Gaba’s lab, A. Dombrovsky’s lab, and S. Manulis-Sasson’s lab, for some of the equipment needed for the experiments.
References
- Chutkan, H., Macdonald, I., Manning, A. and Kuehn, M. J. (2013). Quantitative and qualitative preparations of bacterial outer membrane vesicles. Methods Mol Biol 966: 259-272.
- Goel, A. K., Rajagopal, L. and Sonti, R. V. (2001). Pigment and virulence deficiencies associated with mutations in the aroE gene of Xanthomonas oryzae pv. oryzae. Appl Environ Microbiol 67(1): 245-250.
- Munhoz, C. F., Weiss, B., Hanai, L. R., Zucchi, M. I., Fungaro, M. H., Oliveira, A. L., Monteiro-Vitorello, C. B. and Vieira, M. L. (2011). Genetic diversity and a PCR-based method for Xanthomonas axonopodis detection in passion fruit. Phytopathology 101(4): 416-424.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Mordukhovich, G. and Bahar, O. (2017). Isolation of Outer Membrane Vesicles from Phytopathogenic Xanthomonas campestris pv. campestris. Bio-protocol 7(5): e2160. DOI: 10.21769/BioProtoc.2160.
Category
Microbiology > Microbial cell biology > Organelle isolation
Microbiology > Microbe-host interactions > Bacterium
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










