- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Ca2+ Measurements in Mammalian Cells with Aequorin-based Probes
Published: Vol 7, Iss 5, Mar 5, 2017 DOI: 10.21769/BioProtoc.2155 Views: 8619
Reviewed by: Nicoletta CordaniPia GiovannelliHsin-Yi Chang

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Targeting the Expression of Long Noncoding RNAs in Murine Satellite Cells from Single Myofibers
Martina Macino [...] Chiara Mozzetta
Nov 5, 2021 3631 Views

Preparation and Cultivation of Colonic and Small Intestinal Murine Organoids Including Analysis of Gene Expression and Organoid Viability
Luisa Klemke [...] Ramona Schulz-Heddergott
Jan 20, 2022 7472 Views

Analysis and Quantification of the Mitochondrial–ER Lipidome
Alexis Diaz-Vegas [...] James G. Burchfield
Jul 5, 2024 2587 Views
Abstract
Aequorin is a Ca2+ sensitive photoprotein suitable to measure intracellular Ca2+ transients in mammalian cells. Thanks to recombinant cDNAs expression, aequorin can be specifically targeted to various subcellular compartments, thus allowing an accurate measurement of Ca2+ uptake and release of different intracellular organelles. Here, we describe how to use this probe to measure cytosolic Ca2+ levels and mitochondrial Ca2+ uptake in mammalian cells.
Keywords: Ca2+Background
Aequorin is a 21 kDa photoprotein isolated from jellyfish Aequorea victoria that emits blue light in the presence of Ca2+. In its active form the photoprotein includes an apoprotein and a covalently bound prosthetic group, called coelenterazine. The apoprotein contains four helix-loop-helix ‘EF-hand’ domains, three of which are Ca2+-binding sites. These domains confer to the protein a particular globular structure forming the hydrophobic core cavity that accommodates the coelenterazine. When Ca2+ ions bind to the three high affinity EF-hand sites, coelenterazine is irreversibly oxidized to coelenteramide, with a concomitant release of CO2 and emission of light (Head et al., 2000).
Aequorin began to be widely used when the cDNA encoding the photoprotein was cloned, thus opening the way to recombinant expression. In particular, recombinant aequorin can be expressed not only in the cytoplasm, but also in single intracellular compartments by including specific targeting sequences in the engineered cDNAs (Hartl et al., 1989). To expand the range of Ca2+ sensitivity that can be monitored, point mutations in the EF-hand motives that lower the affinity for Ca2+ have been introduced (Granatiero et al., 2014a and 2014b). Reconstitution of an active recombinant aequorin in living cells is obtained by simple addition of coelenterazine into the medium. Coelenterazine is highly hydrophobic and permeates cell membranes of various cell types. Different coelenterazine analogues have been synthetized and are now commercially available.
Materials and Reagents
- Round glass coverslips 12 mm (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 1014355112NR15 ) - sterile upon autoclave cycle
- 24-well plate (Corning, Costar®, catalog number: 3524 )
- MDA-MB-231 cell line
- MDA-MB-468 cell line
- BT-549 cell line
- Aequorin-expressing plasmids (Brini, 2008)
- Gelatin
- Collagen
- Coelenterazine 0.5 mM stock solution, in methanol (IS Chemical Technology, catalog number: I14-2266 ) - aliquot in 50 μl aliquots. Store at -80 °C. Save it from light
- Agonist (e.g., ATP, Histamine, Bradykinin, Caffeine, Carbachol, Glutamate)
Note: In this protocol example 100 µM ATP is used.
- Milli-Q water
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S9888 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P5405 )
- Magnesium chloride hexahydrate (MgCl2·6H2O) (Sigma-Aldrich, catalog number: M2670 )
- Potassium dihydrogen phosphate (KH2PO4) (Sigma-Aldrich, catalog number: NIST200B )
- Magnesium sulfate heptahydrate (MgSO4·7H2O) (Sigma-Aldrich, catalog number: M2773 )
- HEPES (Sigma-Aldrich, catalog number: H3375 )
- Sodium hydroxide (NaOH) (Sigma-Aldrich, catalog number: 71687 )
- Calcium chloride (CaCl2) (Sigma-Aldrich, catalog number: 449709 )
- Glucose (Sigma-Aldrich, catalog number: G8270 )
- Digitonin (Sigma-Aldrich, catalog number: D5628 or D141 )
- Krebs-Ringer modified buffer (KRB) (see Recipes)
- Digitonin lysis solution (see Recipes)
Equipment
- Windows-based computer
- Perfusion chamber (Elettrofor)
- Low noise photomultiplier (Hamamatsu Photonics K. K., model: H7360-01)
- Peristaltic pump (Gilson’s MINIPULS® 3)
- Water bath (temperature-controlled)
- Photon-counting unit (Hamamatsu Photonics K. K., model: C8855-01 )
Note: Equipment is assembled as depicted in Figure 1.
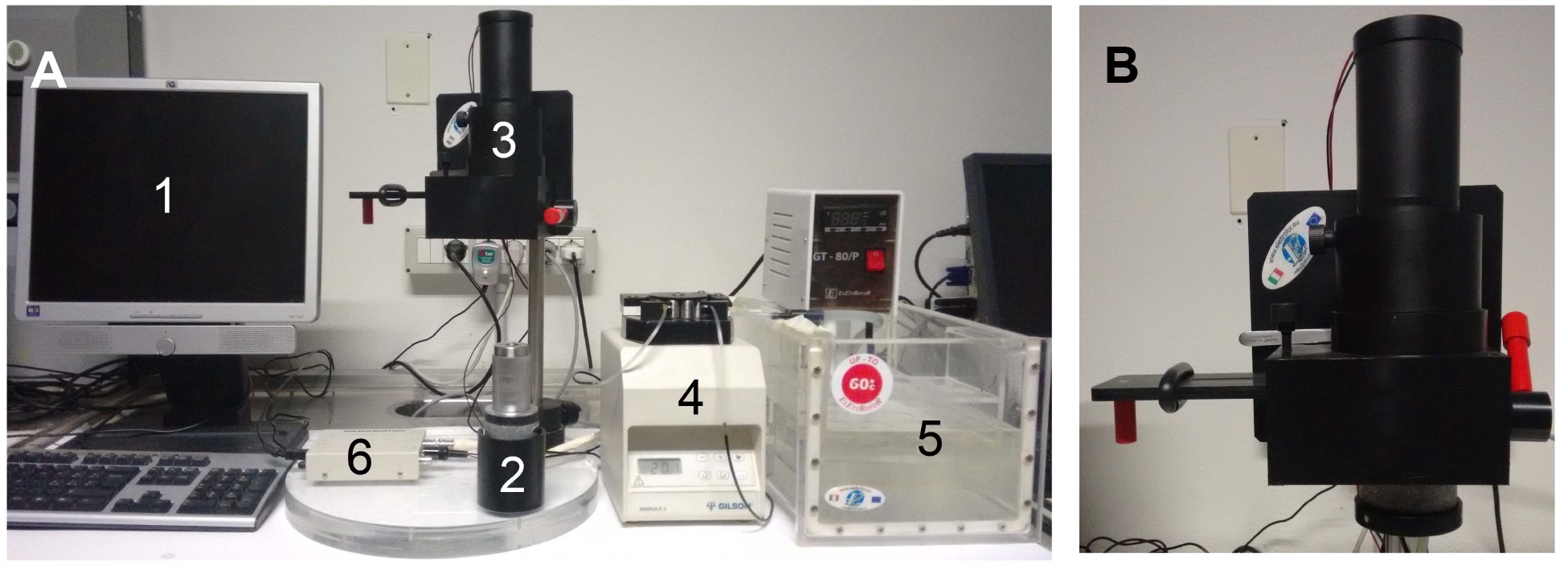
Figure 1. Equipment. A. Fully equipped aequorinometer is composed of: (1) computer, (2) perfusion chamber, (3) photomultiplier, (4) peristaltic pump, (5) water bath, (6) photon-counting unit. B. During the experiment, the perfusion chamber is placed in close proximity to the photomultiplier, protected from light.
Procedure
- Day 1
Plate cells on 12 mm round glass coverslips, in a 24-well plate at 30-50% confluence (about 70,000 cells/well) and let them grow in their specific medium (Figure 2). Pre-treatment of coverslips with gelatin, collagen or poly-lysine to increase cell adherence is not mandatory, but could be recommended for specific cell types.
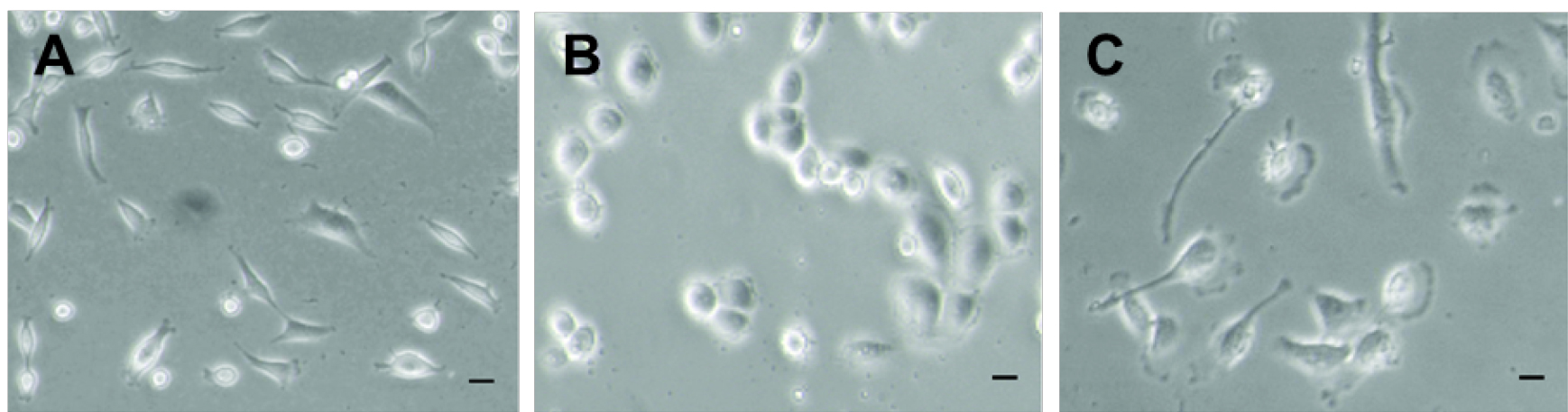
Figure 2. Different mammalian cell lines. MDA-MB-231 (A), MDA-MB-468 (B) and BT-549 (C) breast cancer cell lines at around 40% confluence.
- Day 2
Transfect cells with the proper aequorin-expressing plasmids, according to the expected free [Ca2+] present in the subcellular compartment (Brini et al., 1995; Brini, 2008). Choose the best transfection/infection protocol depending on the targeted cell type. In this example protocol, Aequorin-wt and mitochondrial mutated aequorin (Asp119Ala) are used.
- Day 3
- 24 h after transfection remove medium from the cell plate.
- Wash cells with KRB solution once.
- Incubate cells at 37 °C for 90 min with 5 µM coelenterazine in 200 µl KRB solution.
- Transfer a coverslip containing the transfected cells to the perfusion chamber. Fix the perfusion chamber in close proximity to the photomultiplier (2-3 mm distance) (Figure 3).
- Continuously perfuse cells with KRB, at 37 °C in a water bath. Agonists and other drugs should be added to the same solution.
- Switch on the photomultiplier and start recording the light emission, ideally with a time span of 1 sec (the output of the amplifier-discriminator is captured by the photon-counting unit connected to a Windows-based computer).
- As soon as the background values are stable, stimulate cells with an appropriate agonist concentration, in order to induce Ca2+ release from the endoplasmic reticulum (ER).
- Follow light emission until suppression of Ca2+ signals.
- Terminate the experiment by lysing cells with the digitonin solution, thus discharging the remaining aequorin pool.
- Proceed with data analysis and calibration, as described below.
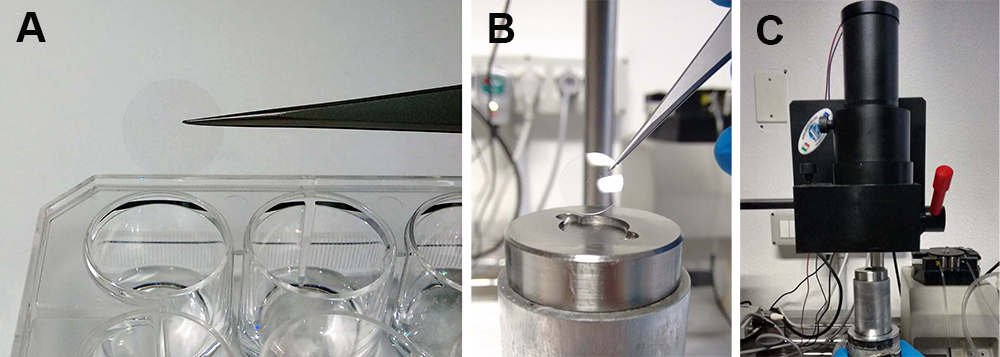
Figure 3. Experimental procedure. The coverslip coated with transfected cells is moved from the plate (A) to the perfusion chamber (B). The perfusion chamber is then placed near the photomultiplier (C).
- 24 h after transfection remove medium from the cell plate.
Data analysis
In order to convert the luminescence signal in [Ca2+], an algorithm has been developed. Three variables are required:
- The total amount of luminescence that each sample would emit in presence of saturating [Ca2+]. In order to calculate this parameter, at the end of each experiment cells are lysed with a digitonin-containing solution (as reported in the Procedure section), and the luminescence emitted by the residual aequorin is measured. Since the total amount of luminescence is directly dependent on the amount of aequorin, this parameter takes into account differences in transfection efficiency.
- Lmax, i.e., the luminescence that would be emitted at a certain time point if all the aequorin had been suddenly discharged. Since aequorin is being constantly consumed, Lmax progressively decreases during the experiment. At each time point, Lmax is calculated by subtracting the luminescence recorded before that point from the total light output of the whole experiment.
- L, i.e., the rate of photon emission at any instant during the experiment.
The [Ca2+] at each time point is a function of L/Lmax (Figure 4). The presence of three Ca2+ binding sites in the aequorin molecule is responsible for the steep relationship between photon emission rate and free [Ca2+]. The rate of luminescence is independent of [Ca2+] at very high (> 10-4 M) and very low [Ca2+] (< 10-7 M). However, it is possible to expand the range of [Ca2+] that can be monitored by choosing the proper recombinant aequorin construct (Figure 5).
Data should be plotted as average of Ca2+max peaks from replicates of each condition. At least 6 replicates per condition are recommended. In addition, a representative Ca2+ trace for each group of samples should be shown.
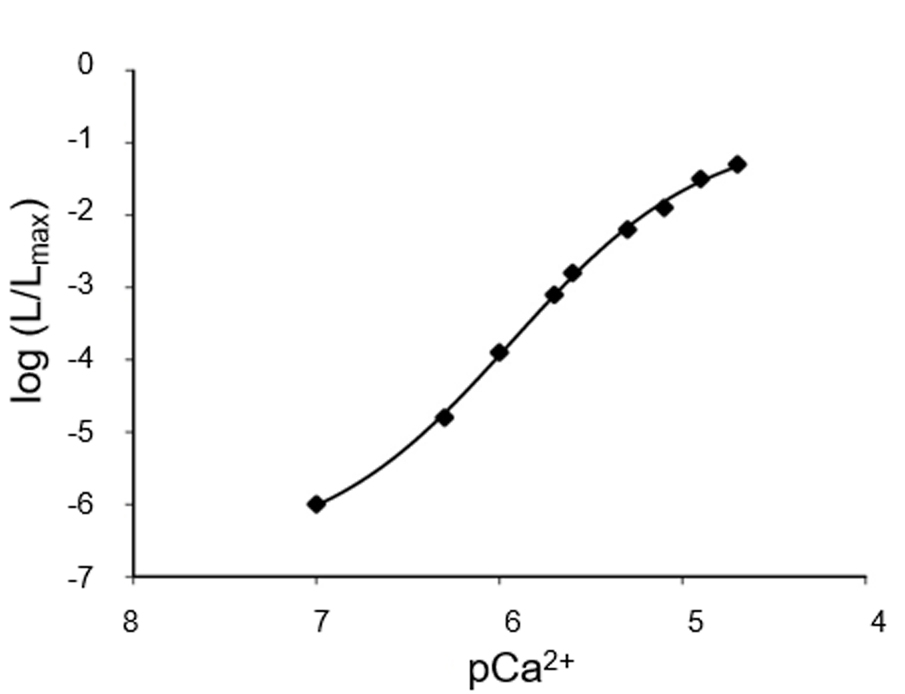
Figure 4. Representative Ca2+ calibration curve. At the end of the experiment, after cell lysis, the total amount of aequorin can be estimated and L/Lmax can be calculated for each data point, thus allowing the conversion of light emitted into free Ca2+ concentration values. N = 6.
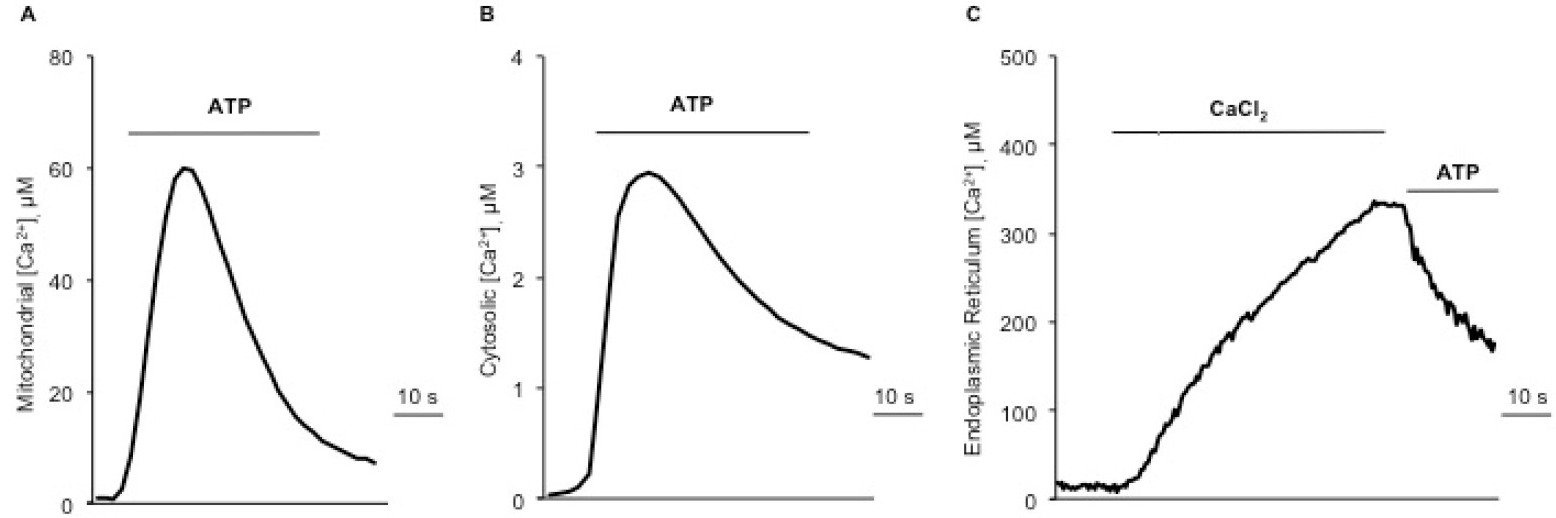
Figure 5. Representative traces of intracellular Ca2+ measurements. A. Mitochondrial Ca2+ uptake upon 100 µM ATP stimulation, using a mutated isoform (Asp119Ala) of aequorin. B. Cytosolic Ca2+ transients upon ATP stimulation, using the wild-type aequorin. C. ER Ca2+ uptake monitored by adding 1 mM CaCl2 to a Ca2+-free KRB, and subsequent Ca2+ release induced by agonist stimulation (ATP).
Notes
- Although aequorin luminescence is influenced neither by K+ nor Mg2+ (which are the most abundant cations in the intracellular environment and thus the most likely source of interference in physiological experiments) both ions are competitive inhibitors of Ca2+-activated luminescence. pH also affects aequorin luminescence at values below 7. For these reasons, experiments with aequorin need to be done in well-controlled conditions of pH and ionic concentrations, notably of Mg2+.
- To stimulate Ca2+ release from ER, the appropriate agonist should be selected for each cell type (e.g., ATP, Histamine, Bradykinin, Caffeine, Carbachol, Glutamate). The choice depends on cell-type specific expression of the G protein-coupled plasma membrane receptors.
Recipes
- Krebs-Ringer modified buffer (KRB)
135 mM NaCl
5 mM KCl
1 mM MgCl2·6H2O
0.4 mM KH2PO4
1 mM MgSO4·7H2O
20 mM HEPES
Adjust the pH to 7.4 with NaOH
Store at 4 °C
Add 1 mM CaCl2 and 5.5 mM glucose, fresh before use
- Digitonin lysis solution
100 μM digitonin
10 mM CaCl2
Store at 4 °C for maximum 4 days
Acknowledgments
The research is supported by grants from the European Union (ERC mitoCalcium, No. 294777 to R.R.); Italian Ministries of Education, University and Research (PRIN to R.R., FIRB to R.R., FIRB Futuro in Ricerca RBFR10EGVP_002 to C.M.); NIH (grant 1P01AG025532-01A1 to R.R.) and French Muscular Dystrophy Association AFM-Téléthon (18857 to C.M.).
References
- Blinks, J. R. (1989). Use of calcium-regulated photoproteins as intracellular Ca2+ indicators. Methods Enzymol 172(4): 164-203.
- Brini, M. (2008). Calcium-sensitive photoproteins. Methods 46(3): 160-166.
- Brini, M., Marsault, R., Bastianutto, C., Alvarez, J., Pozzan, T. and Rizzuto, R. (1995). Transfected aequorin in the measurement of cytosolic Ca2+ concentration ([Ca2+]c). A critical evaluation. J Biol Chem 270(17): 9896-9903.
- Granatiero, V., Patron, M., Tosatto, A., Merli, G. and Rizzuto, R. (2014a). Using targeted variants of aequorin to measure Ca2+ levels in intracellular organelles. Cold Spring Harb Protoc 2014(1): pdb-prot072843.
- Granatiero, V., Patron, M., Tosatto, A., Merli, G. and Rizzuto, R. (2014b). The use of aequorin and its variants for Ca2+ measurements. Cold Spring Harb Protoc 2014(1): pdb-top066118.
- Hartl, F. U., Pfanner, N., Nicholson, D. W. and Neupert, W. (1989). Mitochondrial protein import. Biochim Biophys Acta 988(1): 1-45.
- Head, J. F., Inouye, S., Teranishi, K. and Shimomura, O. (2000). The crystal structure of the photoprotein aequorin at 2.3 Å resolution. Nature 405(6784): 372-376.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Tosatto, A., Rizzuto, R. and Mammucari, C. (2017). Ca2+ Measurements in Mammalian Cells with Aequorin-based Probes. Bio-protocol 7(5): e2155. DOI: 10.21769/BioProtoc.2155.
Category
Cancer Biology > Invasion & metastasis > Cancer therapy
Cell Biology > Organelle isolation > Mitochondria
Molecular Biology > RNA > Transfection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









