- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Adhesion Assay for Murine Bone Marrow Hematopoietic Stem Cells
Published: Vol 7, Iss 4, Feb 20, 2017 DOI: 10.21769/BioProtoc.2135 Views: 12341
Reviewed by: Xiujun FanVivien Jane Coulson-ThomasAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation and Ex Vivo Testing of CD8+ T-Cell Division and Activation Using Mouse Splenocytes
Melissa Dolan [...] John M.L. Ebos
Aug 20, 2025 3878 Views

Detection of Autophagy in Human Peripheral Blood Mononuclear Cells Using Guava® Autophagy and Flow Cytometry
Melanie Scherer [...] Jörg Bergemann
Sep 20, 2025 1418 Views

Protocol for the Isolation and Analysis of Extracellular Vesicles From Peripheral Blood: Red Cell, Endothelial, and Platelet-Derived Extracellular Vesicles
Bhawani Yasassri Alvitigala [...] Lallindra Viranjan Gooneratne
Nov 5, 2025 1448 Views
Abstract
Hematopoietic stem cells (HSCs) are defined by their functional abilities to self-renew and to give rise to all mature blood and immune cell types throughout life. Most HSCs are retained in a non-motile quiescent state within a specialized protective microenvironment in the bone marrow (BM) termed the niche. HSCs are typically distinguished from other adult stem cells by their motility capacity. Movement of HSCs across the physical barrier of the marrow extracellular matrix and blood vessel endothelial cells is facilitated by suppression of adhesion interactions, which are essential to preserve the stem cells retained within their BM niches. Importantly, homing of HSCs to the BM following clinical transplantation is a crucial first step for the repopulation of ablated BM as in the case of curative treatment strategies for hematologic malignancies. The homing process ends with selective access and anchorage of HSCs to their specialized niches within the BM. Adhesion molecules are targets to either enhance homing in cases of stem cell transplantation or reduce BM retention to harvest mobilized HSCs from the blood of matched donors. A major adhesion protein which is functionally expressed on HSCs and is involved in their homing and retention is the integrin alpha4beta1 (Very late antigen-4; VLA4). In this protocol we introduce an adhesion assay optimized for VLA4 expressing murine bone marrow stem cells. This assay quantifies adherent HSCs by flow cytometry with HSC enriching cell surface markers subsequent to the isolation of VLA4 expressing adherent cells.
Keywords: Very late antigen 4Background
HSCs are mostly retained in the BM and are regulated by adhesive interactions with their microenvironment, the niche. In this way HSCs are kept in a non-motile quiescent state which protects them from DNA damaging agents (Boulais and Frenette, 2015; Mendelson and Frenette, 2014; Miyamoto et al., 2011; Morrison and Scadden, 2014). The defining properties of HSCs are their functional ability to durably repopulate the irradiated BM of transplanted recipients, which requires their homing, self-renewal and developmental potential (Gur-Cohen et al., 2016). Since adhesion gives rise to activation of intracellular signaling pathways, the type of interaction can mirror the developmental state and behavior of the cells (Sugiyama et al., 2006). Adhesion assays are methods to distinguish between adhesive and non-adhesive cells. In this protocol we introduce a cell adhesion assay under static conditions that separates VLA4 expressing adhesive cells from non-adhesive cells, which are quantified by FACS analysis.
In mouse, hematopoietic stem and progenitor cells (HSPCs) are enriched in a population that lacks lineage markers (Lin; CD8a, CD4, GR1, B220, TER-119, CD11b), and expresses c-Kit (K) and Sca-1 (S). Hence, these cells are also called Lin- Sca-1+ c-Kit+ (LSK) cells (Adolfsson et al., 2001; Okada et al., 1991; Spangrude et al., 1988). EPCR (endothelial protein C receptor) has been identified as a stem cell marker also in various other tissues (Balazs et al., 2006; Iwasaki et al., 2010; Kent et al., 2009; Ramalho-Santos et al., 2002; Wang and Gerdes, 2015).
Adhesion molecules play a major role in the retention and egress of these HSCs in the BM and to the blood circulation. VLA4 is a receptor for both fibronectin and VCAM-1 and is expressed by most leukocytes, as well as by some non-hematopoietic cells (Hemler et al., 1990), while its expression is higher on murine BM EPCR+ LT-HSCs as compared to EPCR negative progenitor cells and circulating LT-HSC (Gur-Cohen et al., 2015). It has long been proposed that VLA4 expression by LT-HSCs might be important for binding and detachment of stem cells within the human BM microenvironment. Inhibition of VLA4 or VCAM-1 binding by neutralizing antibodies causes mobilization of HSPCs from the BM to the blood circulation of mice and primates (Craddock et al., 1997; Papayannopoulou et al., 1995) which is consistent with the notion that VLA4 is crucial for CXCL12/CXCR4-mediated LT-HSC quiescent retention in the BM (Papayannopoulou et al., 1995; Papayannopoulou and Scadden, 2008). In addition to HSPC BM retention, VLA4 is also essential for murine HSPC BM homing (Papayannopoulou and Craddock, 1997). VLA4 possesses different conformations that correlate with its affinity states (Alon et al., 1995; Chen et al., 1999; Feigelson et al., 2001) which are influenced by divalent cations and inside-out signaling (Chigaev et al., 2003; Chigaev et al., 2011). The majority of VLA4 affinity inside- out signaling is mediated by G-protein coupled receptors (Laudanna et al., 2002; Chigaev et al., 2008; Arnaout et al., 2007). Furthermore, elevation of intracellular nitric oxide (NO) was shown to cause cGMP-mediated inhibition of VLA4 affinity (Chigaev et al., 2011). We have previously shown two different pathways, the aPC-EPCR-PAR1 and the thrombin-PAR1 axis, which regulate the NO level up and down, respectively. Thereby, these pathways influence a number of intracellular molecules including Cdc42, CXCR4 and VLA4 leading to retention or mobilization of HSPCs (Gur-Cohen et al., 2015). As described by Gur-Cohen et al. (2015), we herein propose the VLA4 mediated adhesion assay for EPCR+ stem cells as a powerful tool to predict LT-HSC retention potential to their bone marrow niches.
Materials and Reagents
- Tissue culture 6-well plates (Corning, Costar®, catalog number: 3516 )
- NuncTM 8.8 cm2 Petri dish (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 153066 )
- 1 ml slip tip Sub-Q syringe with disposable 26 G x 5/8 inch needle (BD, catalog number: 309597 )
- Autoclaved pipet tips:
- FACS tubes (Corning, Falcon®, model: 352054 )
- Sterilized 40 µm pore-sized nylon mesh (Sinun Tech, catalog number: Plymer Screends ) (see Recipes)
- Mice
- Fibronectin (Sigma-Aldrich, catalog number: F0895 )
- Human CXCL12 (human SDF-1 alpha) (reprokineTM, catalog number: RKP48061 ) or (PeproTech, Rocky Hill, NJ, USA)
- BSA (Sigma-Aldrich, catalog number: A9647 )
- LymphoprepTM Ficoll (STEMCELL Technologies, catalog number: 07861 )
- Antibodies:
- EPCR PE (Affymetrix, eBioscience, catalog number: 12-2012-82 ) or EPCR PerCP-eFluor 710(Affymetrix, eBioscience, catalog number: 46-2012-80 ) or EPCR Biotin (Affymetrix, eBioscience, catalog number: 13-2012-82 ) combined with streptavidin-PE (BioLegend, catalog number: 405203 )
- Sca-1 PE (BioLegend, catalog number: 108108 ) or Sca-1 PE/Cy7 (BioLegend, catalog number: 108114
- c-Kit APC (BioLegend, catalog number: 105812 )
- Lineage antibodies
Note: This is a set of antibodies that target the antigens i-vi listed below which are cell lineage markers. In mice these markers do not occur on stem and progenitor cells.- CD8a FITC (BioLegend, catalog number: 100706 )
- CD4 FITC (BioLegend, catalog number: 100406 )
- GR1 FITC (BioLegend, catalog number: 108406 )
- B220 FITC (BioLegend, catalog number: 103206 )
- TER-119 FITC (BioLegend, catalog number: 116206 )
- CD11b FITC(BioLegend, catalog number: 101206 )
- Or Lineage Cocktail-Biotin (Miltenyi Biotec, catalog number: 130-092-613 ) combined with streptavidin-FITC (BioLegend; catalog number: 405202 )
Note: The combination of fluorophores used for the experiment needs to be adjusted according to the experimental demands.
- CD8a FITC (BioLegend, catalog number: 100706 )
- EPCR PE (Affymetrix, eBioscience, catalog number: 12-2012-82 ) or EPCR PerCP-eFluor 710(Affymetrix, eBioscience, catalog number: 46-2012-80 ) or EPCR Biotin (Affymetrix, eBioscience, catalog number: 13-2012-82 ) combined with streptavidin-PE (BioLegend, catalog number: 405203 )
- 1x Dulbecco’s phosphate buffered saline without calcium and magnesium (PBS-/-) (generated from 10x PBS, see Recipes) (Biological Industries, catalog number: 02-023-5A )
- Roswell park memorial institute (RPMI) 1640 medium ([+] 300 mg/L L-glutamine, [+] 25 mM HEPES) (Thermo Fisher Scientific, GibcoTM, catalog number: 52400025 )
- Fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 12657-029 )
- Penicillin-streptomycin (Pen-Strep) solution (Biological Industries, catalog number: 03-031-1B )
- L-glutamine solution (Biological Industries, catalog number: 03-020-1B )
- Gentian violet (Sigma-Aldrich, catalog number: G2039 )
- Acetic acid (Sigma-Aldrich, catalog number: ARK2183 )
- 0.1 M sodium azide solution (Sigma-Aldrich, catalog number: 0 8591 )
- Acetone (Sigma-Aldrich, catalog number: 40289 )
Note: This product has been discontinued. - Ethanol (Sigma-Aldrich, catalog number: 24103 )
Note: This product has been discontinued. - Cell dissociation solution (Sigma-Aldrich, catalog number: C5914 )
- ddH2O
- Coating solution (see Recipes)
- Blocking solution with 2% (m/v) BSA (see Recipes)
- Complete RPMI medium (see Recipes)
- Turk’s solution (see Recipes)
- FACS buffer (see Recipes)
Equipment
- HeracellTM 150i CO2 incubator (Thermo Fisher Scientific, Thermo ScientificTM, model: 150i CO2 incubator)
- Autoclave
- Scissors
- Forceps
- Refrigerator (4° C)
- Centrifuge (Eppendorf, model: 5810R )
- Centrifuge swing-bucket rotor A-462 4 x 250 ml rectangular buckets (Eppendorf, catalog number: 5810709008 )
- Adapters (Eppendorf, catalog number: 5810752000 )
- Finnpipette model 4500 single channel pipette:
- 0.5-10 μl (Thermo Fisher Scientific, Thermo Scientific, catalog number: FA-10R )
- 5-40 μl (Thermo Fisher Scientific, Thermo Scientific, catalog number: FA-40R )
- 20-200 μl (Thermo Fisher Scientific, Thermo Scientific, catalog number: FA-200R )
- 100-1,000 μl (Thermo Fisher Scientific, Thermo Scientific, catalog number: FA-1000R )
- Inverted light microscope (Olympus, model: CHK2-F-GS )
Note: This product has been discontinued by the manufacturer. - Counting chamber/hemocytometer (Reichert Bright-Line) (Sigma-AIdrich, Bright-LineTM, model: Z359629 )
- Flow cytometer (model optional):
- MACSQuant VYB instrument (Miltenyi Biotec, model: 130096116 )
- FACSCalibur instrument (BD)
- FACS LSRII instrument (with all four fixed-aligned lasers) (BD)
- MACSQuant VYB instrument (Miltenyi Biotec, model: 130096116 )
Software
- CellQuest software
- FACSDiva software
- FlowJo (Tree Star or V10) or MacsQuant
Procedure
- Coating and preparation
- Coat tissue culture 6-well plates with 25 µg/ml fibronectin and 2.5 µg/ml CXCL12 overnight at 4 °C. For the coating, it is recommended to prepare a mix of fibronectin and CXCL12 in PBS-/- (coating medium) and to calculate the amount needed so that each well will be covered with 500 µl coating medium. Fibronectin stock (1 mg/ml) and CXCL12 (50 µg/ml) were stored at -20 °C.
- After overnight incubation, discard the coating medium gently and wash the plates with 1 ml PBS-/- for a few seconds. Repeat washing 3 times.
- Perform blocking with 2% BSA in PBS-/- for 30 min at room temperature. Blocking solution should cover each well of the plates, and therefore it is recommended to use a minimum of 500 µl blocking solution for each well.
- Discard the blocking solution gently and wash plates with 1 ml PBS-/- for a few seconds while tilting the plates slowly. Repeat washing 3 times.
- Plates are ready to be seeded with BM mononuclear cells.
- Coat tissue culture 6-well plates with 25 µg/ml fibronectin and 2.5 µg/ml CXCL12 overnight at 4 °C. For the coating, it is recommended to prepare a mix of fibronectin and CXCL12 in PBS-/- (coating medium) and to calculate the amount needed so that each well will be covered with 500 µl coating medium. Fibronectin stock (1 mg/ml) and CXCL12 (50 µg/ml) were stored at -20 °C.
- Obtaining bone marrow mononuclear cells
- Sacrifice desired mice by CO2 euthanasia or cervical dislocation.
- Extract the femurs, tibias and pelvises using forceps and sharp scissors. Place the bones on a small dish supplemented with PBS-/-.
- Flush total bone marrow cells from the bone cavity with 1-2 ml PBS-/- (per mouse) using a 1 ml syringe and 16 G needle (Figure 1). Obtain single cell suspension by resuspending the cell solution using the same syringe.
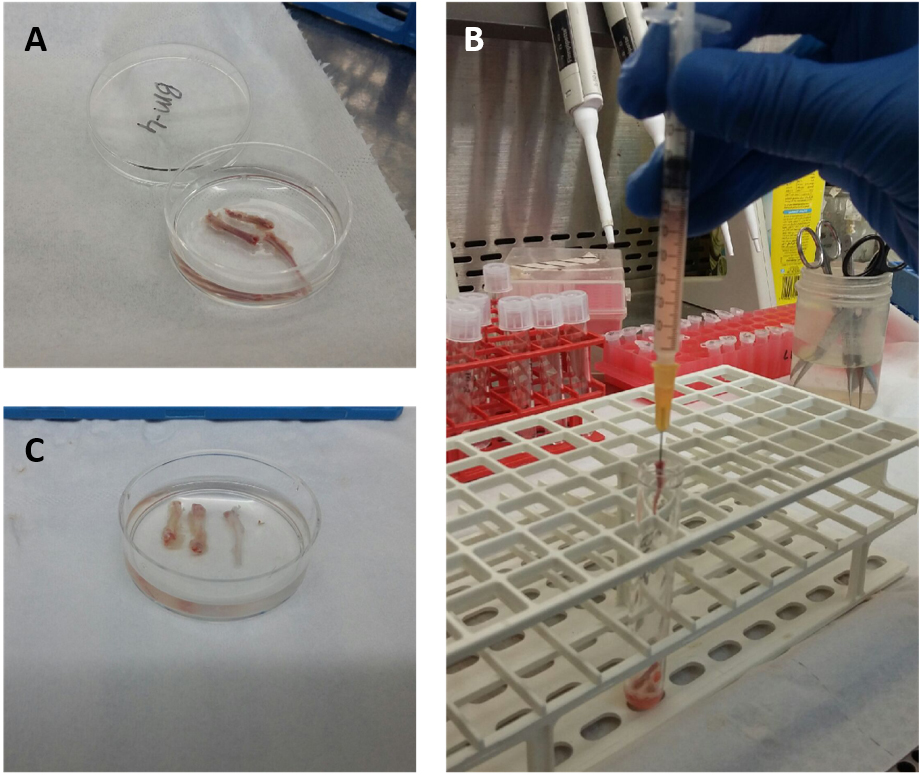
Figure 1. Process of flushing bone marrow out of murine tibia. The red color of the extracted bones indicates the presence of the BM (A). Drilling the needle into the bone and flushing the BM with PBS out of the bone (B) until the bones look white enough (C). - Carefully pipet (very slowly) ficoll to the bottom of each tube with a dilution ratio of 1:2 to the bone marrow cells (i.e., if you have a total of 2 ml bone marrow cells in PBS, pipette 1 ml ficoll to the bottom of the tube) (Figures 2A and 2B).
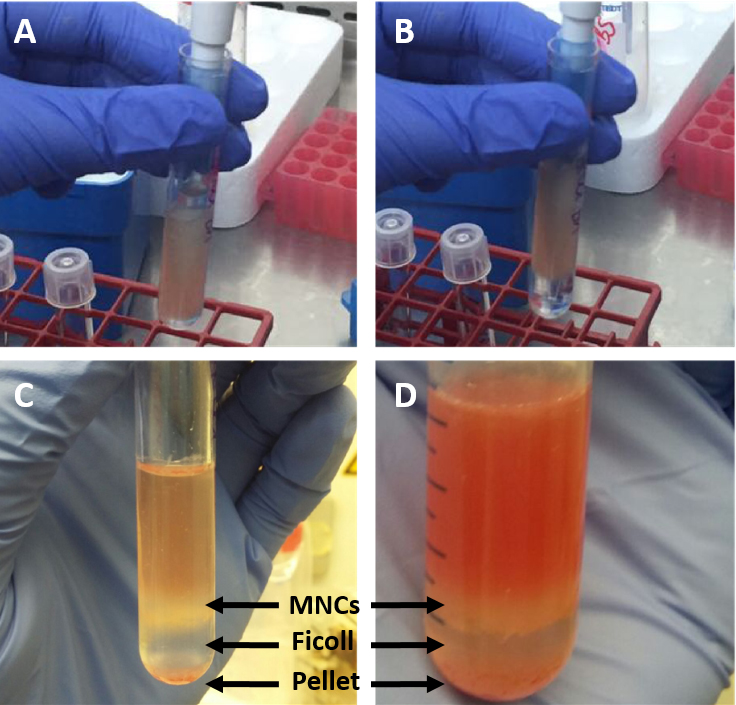
Figure 2. Process of MNC isolation from BM. Ficoll is added to the bottom of the tube underneath the suspended BM (A) forming a lower clear ficoll phase and an upper cell suspension phase (B). After centrifugation the two phases are separated by thin white phase consisting of MNCs (C) which is more prominent the more BM is used for the isolation (D). - Centrifuge the cells at 652 x g for 25 min, without a brake (important) at room temperature (mononuclear cells will not be separated if the centrifuge is cold).
- Carefully remove the tubes from the centrifuge. At the middle phase a fine white ring-like shape should form, containing the bone marrow mononuclear cells (Figures 2C and 2D)
- Carefully, discard the fluids on top of the ring with a pipet, while avoiding touching the mononuclear fraction. Collect the ring of cells with 200 µl pipette tip into a new FACS tube.
- Wash the collected mononuclear cells with 2 ml PBS-/- and centrifuge the cells at 452.8 x g for 5 min.
- Discard the supernatant and resuspend the cell pellet in 1 ml complete RPMI (see Recipes).
- Count the cells by diluting the cells 1:10 with Turk’s solution. Count the cells under the inverted light microscope using a hemocytometer. From one mouse typically one will get 20-40 million mononuclear cells.
- Sacrifice desired mice by CO2 euthanasia or cervical dislocation.
- Adhesion assay (Figure 3)
- Seed bone marrow mononuclear cells at a density of 5 x 106 cells per 1 ml complete RPMI medium.
- Allow the cells to adhere to the coated plates for 2 h at 37 °C in an incubator.
- Collect non-adherent cells to a new FACS tube by gently rinsing each well with complete RPMI medium. Make sure not to touch the bottom and only add RPMI by placing the pipet tip against the wall of each well.
- Gently wash the plates 3 times with PBS-/- by tilting the plates back and forth. Make sure not to touch the bottom and only add PBS-/- by placing the pipet tip against the wall of each well.
- To collect the adherent cells, incubate the plates with cell dissociation buffer for 10 min at 37 °C in the incubator.
- Monitor the cells under the inverted microscope. If cells are still attached to the bottom, resuspend the cells by using pipet and incubate the plate for additional 5-10 min in the presence of the cell dissociation buffer.
- Once the cells are detached from the well, vigorously rinse each well using a pipet and collect the adherent fraction to a new FACS tube. Subsequently, add 1 ml ice cold PBS-/- to each well, rinse thoroughly and collect the fluids into the adherent cell fraction to a different FACS tube.
- Monitor the cells under the inverted microscope. If additional cells remained on the plate, vigorously rinse each well again with ice cold PBS-/- and keep collecting the fluids into the adherent cell fraction.
- Centrifuge the adherent and nonadherent cell fractions at 452.8 x g, for 5 min.
- Discard the supernatant carefully.
- Add 100 µl PBS to each pellet. The cells are now ready to be stained and analyzed.
- Seed bone marrow mononuclear cells at a density of 5 x 106 cells per 1 ml complete RPMI medium.
- FACS staining and analysis (Figure 4)
- Add 1 µl of each lineage antibody (CD8a FITC, CD4 FITC, GR1 FITC, B220 FITC, TER-119 FITC, CD11b FITC) and 2 µl Sca1 PeCy7, cKit APC and EPCR PE antibodies to remaining solution on the bottom of each tube.
Note: Do not forget to include a negative control in order to be able to distinguish between the autofluorescence and positive signal during the flow cytometry. - Resuspend the pellets by vortexing the tubes briefly.
- Leave the samples at 4 °C for 30 min.
Note: If biotin-coupled antibodies are used, resuspend the cells in 1 ml PBS after the incubation at 4 °C and repeat the steps from C9 until this step. At step D1 add 1 µl of FITC-coupled streptavidin to the remaining solution of each tube. - Resuspend each sample in 1 ml FACS buffer and filter through a 40 µm mesh into a new tube to avoid clots during flow cytometry.
- Centrifuge the sample at 452.8 x g for 5 min.
- Discard the supernatant carefully, add 300 µl of FACS buffer to each sample and resuspend the cells by vortexing.
- Read the samples using a flow cytometer. Cell populations are analyzed with a FACSCalibur instrument with CellQuest software, with a MacsQuant instrument or with a FACS LSRII instrument with FACSDiva software. Please consider that the combination of some fluorophores may cause an overlap in signals within the flow cytometer. Therefore, to set good compensation is mandatory.
- Data are analyzed with FlowJo (Tree Star or V10) or MacsQuant according to the gating strategy illustrated in Figure 1 (see Data analysis section as well):
- Gate for live cells using forward (FSC) and sideward scatter (SSC).
- Gate for lineage negative cells using SSC and the channel for the fluorophore coupled to the lineage antibodies (Figure 5).
- Gate for SK cells selecting the c-Kit positive and Sca-1 positive cell population (Figure 5).
- Create a histogram overlay displaying the EPCR signals of the LSK population within the negative control, the adherent and nonadherent fractions.
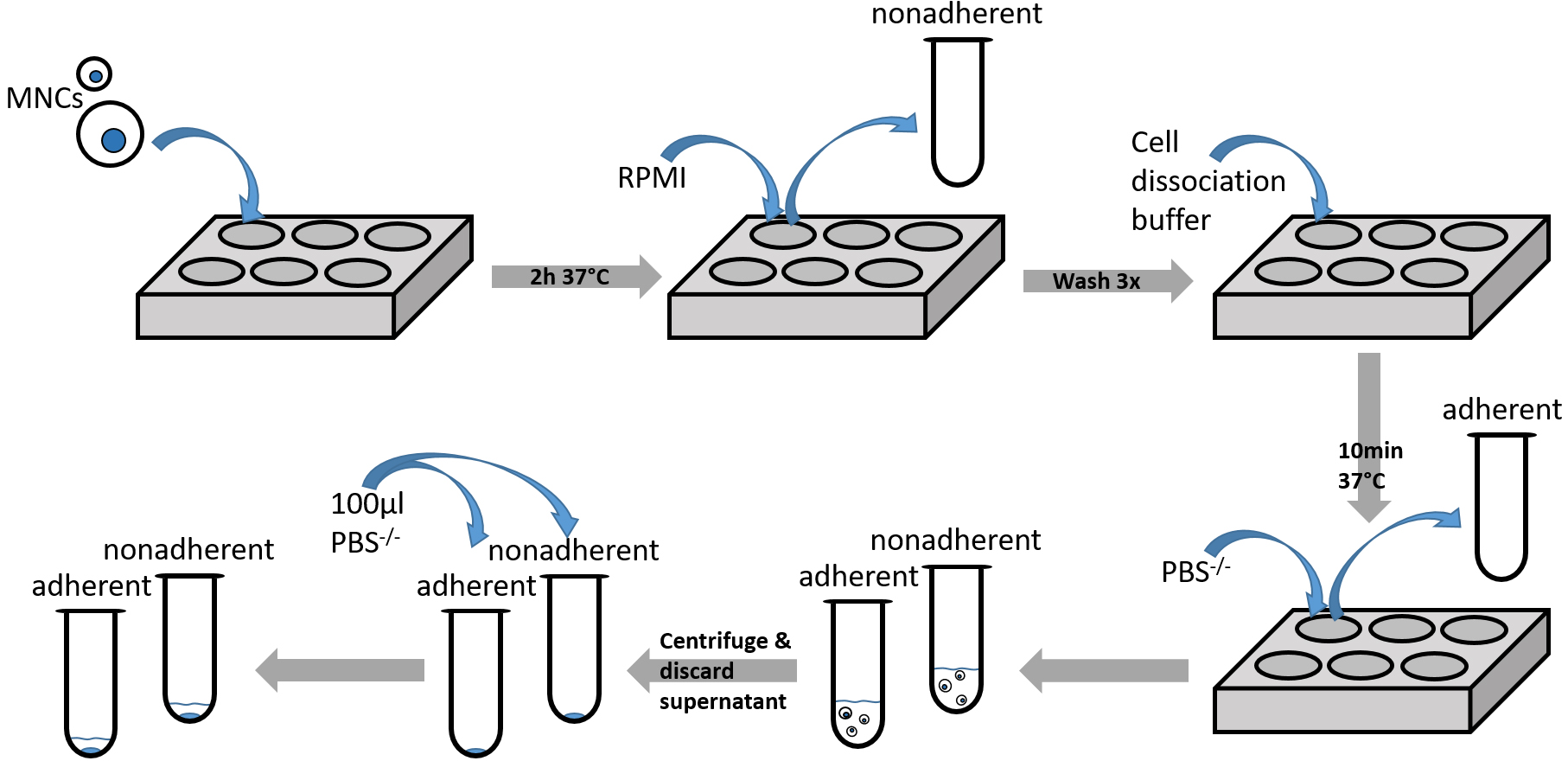
Figure 3. Diagramm showing steps of adhesion assay procedure (Procedure C)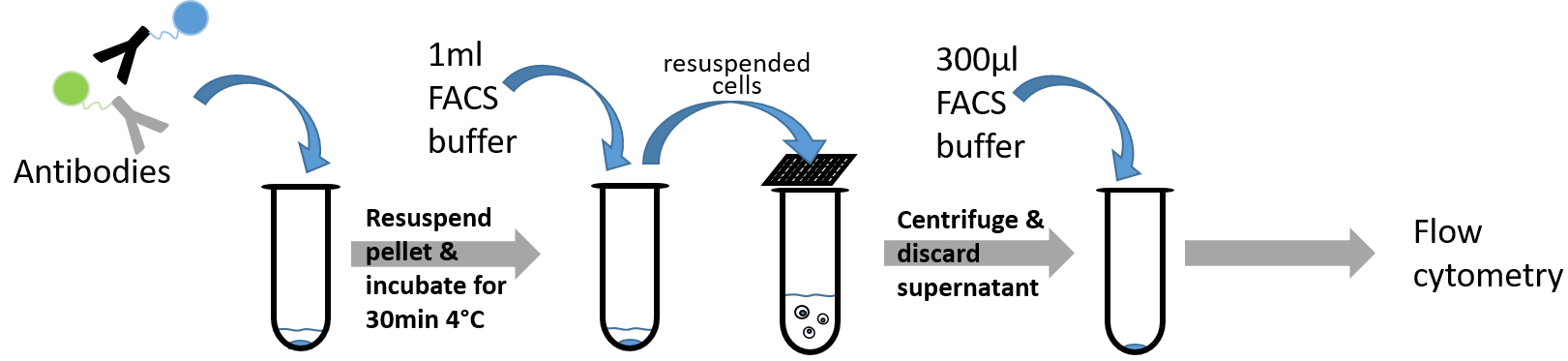
Figure 4. Diagramm showing steps of FACS staining procedure (Procedure D)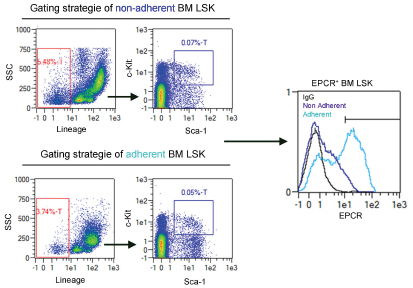
Figure 5. Gating strategy of EPCR positive bone marrow LSK cells (adapted from Supp. Figure 5a of Gur-Cohen et al., 2015)
- Gate for live cells using forward (FSC) and sideward scatter (SSC).
- Add 1 µl of each lineage antibody (CD8a FITC, CD4 FITC, GR1 FITC, B220 FITC, TER-119 FITC, CD11b FITC) and 2 µl Sca1 PeCy7, cKit APC and EPCR PE antibodies to remaining solution on the bottom of each tube.
Data analysis
For further analysis information concerning gating strategies and statistical analysis you can consult the article Gur-Cohen et al. (2015) at the following link: http://www.nature.com/nm/journal/v21/n11/full/nm.3960.html.
Recipes
- 1x PBS-/-
Add 50 ml of 10x PBS to 450 ml of dH2O - Coating solution
1x PBS with 25 µg/ml fibronectin and 2.5 µg/ml CXCL12 - Blocking solution with 2% (m/v) BSA
Add 2 g of BSA per 100 ml 1x PBS - Complete RPMI medium
88% RPMI 1640 medium
10% FBS
1% Pen-Strep solution
1% L-glutamine solution - Turk’s solution
50 mg of gentian violet
5 ml of acetic acid
495 ml of ddH2O
Dilute gentian violet in acetic acid and ddH2O - FACS buffer
Note: Mix this buffer in a plastic bottle.
450 ml ddH2O
18 µl sodium azide
50 ml 10x PBS
5 ml FBS - Sterilized 24 µm pore-sized nylon mesh
- Cut nylon mesh into approx. 3 x 3 pieces
- Incubate the meshes for 20 min at room temperature in acetone under the chemical hood
- Incubate the meshes for 20 min at room temperature in absolute ethanol
- Let the meshes dry overnight
- Sterilize the meshes by autoclaving them at 121 °C for 60 min
Acknowledgments
This study was supported by the Israel Science Foundation (851/13), the Ernest and Bonnie Beutler Research Program of Excellence in Genomic Medicine and FP7-HEALTH-2010 (CELL-PID 261387) (T.L.) and the DKFZ, Germany.
References
- Adolfsson, J., Borge, O. J., Bryder, D., Theilgaard-Mönch, K., Åstrand-Grundström, I., Sitnicka, E. and Jacobsen, S. E. (2001). Upregulation of Flt3 expression within the bone marrow Lin− Sca1+ c-kit+ stem cell compartment is accompanied by loss of self-renewal capacity. Immunity 15(4): 659-669.
- Alon, R., Kassner, P. D., Carr, M. W., Finger, E. B., Hemler, M. E. and Springer, T. A. (1995). The integrin VLA-4 supports tethering and rolling in flow on VCAM-1. J Cell Biol 128(6): 1243-1253.
- Arnaout, M. A., Goodman, S. L. and Xiong, J. P. (2007). Structure and mechanics of integrin-based cell adhesion. Curr Opin Cell Biol 19(5): 495-507.
- Balazs, A. B., Fabian, A. J., Esmon, C. T. and Mulligan, R. C. (2006). Endothelial protein C receptor (CD201) explicitly identifies hematopoietic stem cells in murine bone marrow. Blood 107(6): 2317-2321.
- Boulais, P. E. and Frenette, P. S. (2015). Making sense of hematopoietic stem cell niches. Blood 125(17): 2621-2629
- Chen, L. L., Whitty, A., Lobb, R. R., Adams, S. P. and Pepinsky, R. B. (1999). Multiple activation states of integrin α4β1 detected through their different affinities for a small molecule ligand. J Biol Chem 274(19): 13167-13175.
- Chigaev, A., Smagley, Y. and Sklar, L. A. (2011). Nitric oxide/cGMP pathway signaling actively down-regulates alpha4beta1-integrin affinity: an unexpected mechanism for inducing cell de-adhesion. BMC Immunol 12: 28.
- Chigaev, A., Zwartz, G., Graves, S. W., Dwyer, D. C., Tsuji, H., Foutz, T. D., Edwards, B. S., Prossnitz, E. R., Larson, R. S. and Sklar, L. A. (2003). Alpha4beta1 integrin affinity changes govern cell adhesion. J Biol Chem 278(40): 38174-38182.
- Chigaev, A., Waller, A., Amit, O. and Sklar, L. A. (2008). Galphas-coupled receptor signaling actively down-regulates alpha4beta1-integrin affinity: a possible mechanism for cell de-adhesion. BMC Immunol 9: 26.
- Craddock, C. F., Nakamoto, B., Andrews, R. G., Priestley, G. V. and Papayannopoulou, T. (1997). Antibodies to VLA4 integrin mobilize long-term repopulating cells and augment cytokine-induced mobilization in primates and mice. Blood 90(12): 4779-4788.
- Feigelson, S. W., Grabovsky, V., Winter, E., Chen, L. L., Pepinsky, R. B., Yednock, T., Yablonski, D., Lobb, R. and Alon, R. (2001). The Src kinase p56(lck) up-regulates VLA-4 integrin affinity. Implications for rapid spontaneous and chemokine-triggered T cell adhesion to VCAM-1 and fibronectin. J Biol Chem 276(17): 13891-13901.
- Gur-Cohen, S., Itkin, T., Chakrabarty, S., Graf, C., Kollet, O., Ludin, A., Golan, K., Kalinkovich, A., Ledergor, G., Wong, E., Niemeyer, E., Porat, Z., Erez, A., Sagi, I., Esmon, C. T., Ruf, W. and Lapidot, T. (2015). PAR1 signaling regulates the retention and recruitment of EPCR-expressing bone marrow hematopoietic stem cells. Nat Med 21(11): 1307-1317.
- Gur-Cohen, S., Kollet, O., Graf, C., Esmon, C. T., Ruf, W. and Lapidot, T. (2016). Regulation of long-term repopulating hematopoietic stem cells by EPCR/PAR1 signaling. Ann N Y Acad Sci 1370(1): 65-81.
- Hemler, M. E., Elices, M. J., Parker, C. and Takada, Y. (1990). Structure of the integrin VLA-4 and its cell-cell and cell-matrix adhesion functions. Immunol Rev 114: 45-65.
- Laudanna, C., Kim, J. Y., Constantin, G. and Butcher, E. (2002). Rapid leukocyte integrin activation by chemokines. Immunol Rev 186: 37-46.
- Iwasaki, H., Arai, F., Kubota, Y., Dahl, M. and Suda, T. (2010). Endothelial protein C receptor-expressing hematopoietic stem cells reside in the perisinusoidal niche in fetal liver. Blood 116(4): 544-553.
- Kent, D. G., Copley, M. R., Benz, C., Wohrer, S., Dykstra, B. J., Ma, E., Cheyne, J., Zhao, Y., Bowie, M. B., Zhao, Y., Gasparetto, M., Delaney, A., Smith, C., Marra, M. and Eaves, C. J. (2009). Prospective isolation and molecular characterization of hematopoietic stem cells with durable self-renewal potential. Blood 113(25): 6342-6350.
- Mendelson, A. and Frenette, P. S. (2014). Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat Med 20(8): 833-846.
- Miyamoto, K., Yoshida, S., Kawasumi, M., Hashimoto, K., Kimura, T., Sato, Y., Kobayashi, T., Miyauchi, Y., Hoshi, H., Iwasaki, R., Miyamoto, H., Hao, W., Morioka, H., Chiba, K., Kobayashi, T., Yasuda, H., Penninger, J. M., Toyama, Y., Suda, T. and Miyamoto, T. (2011). Osteoclasts are dispensable for hematopoietic stem cell maintenance and mobilization. J Exp Med 208(11): 2175-2181.
- Morrison, S. J. and Scadden, D. T. (2014). The bone marrow niche for haematopoietic stem cells. Nature 505(7483): 327-334.
- Okada, S., Nakauchi, H., Nagayoshi, K., Nishikawa, S., Nishikawa, S., Miura, Y. and Suda, T. (1991). Enrichment and characterization of murine hematopoietic stem cells that express c-kit molecule. Blood 78(7): 1706-1712.
- Papayannopoulou, T. and Craddock, C. (1997). Homing and trafficking of hemopoietic progenitor cells. Acta Haematol 97(1-2): 97-104.
- Papayannopoulou, T., Craddock, C., Nakamoto, B., Priestley, G. V. and Wolf, N. S. (1995). The VLA4/VCAM-1 adhesion pathway defines contrasting mechanisms of lodgement of transplanted murine hemopoietic progenitors between bone marrow and spleen. Proc Natl Acad Sci U S A 92(21): 9647-9651.
- Papayannopoulou, T. and Scadden, D. T. (2008). Stem-cell ecology and stem cells in motion. Blood 111(8): 3923-3930.
- Ramalho-Santos, M., Yoon, S., Matsuzaki, Y., Mulligan, R. C. and Melton, D. A. (2002). "Stemness": transcriptional profiling of embryonic and adult stem cells. Science 298(5593): 597-600.
- Spangrude, G. J., Heimfeld, S. and Weissman, I. L. (1988). Purification and characterization of mouse hematopoietic stem cells. Science 241(4861): 58-62.
- Sugiyama, T., Kohara, H., Noda, M. and Nagasawa, T. (2006). Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity 25(6): 977-988.
- Wang, X. and Gerdes, H. H. (2015). Transfer of mitochondria via tunneling nanotubes rescues apoptotic PC12 cells. Cell Death Differ 22(7): 1181-1191.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Avci, S., Gur-Cohen, S., Avemaria, F. and Lapidot, T. (2017). Adhesion Assay for Murine Bone Marrow Hematopoietic Stem Cells. Bio-protocol 7(4): e2135. DOI: 10.21769/BioProtoc.2135.
Category
Stem Cell > Adult stem cell > Hematopoietic stem cell
Cell Biology > Cell movement > Cell adhesion
Cell Biology > Cell-based analysis > Flow cytometry
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









