- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Analysis of the Virulence of Uropathogenic Escherichia coli Strain CFT073 in the Murine Urinary Tract
Published: Vol 7, Iss 3, Feb 5, 2017 DOI: 10.21769/BioProtoc.2129 Views: 11520
Reviewed by: Andrea PuharMigla MiskinyteAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In vivo Electroporation of Skeletal Muscle Fibers in Mice
Steven J. Foltz [...] Hyojung J. Choo
Jul 5, 2023 1829 Views

Cochlear Organ Dissection, Immunostaining, and Confocal Imaging in Mice
Chenyu Chen [...] Dongdong Ren
Jan 20, 2025 3773 Views

Isolation and Imaging of Microvessels From Brain Tissue
Josephine K. Buff [...] Sophia M. Shi
Aug 5, 2025 2636 Views
Abstract
This urinary tract infection model was used to monitor the efficacy of a new virulence factor of the uropathogenic Escherichia coli strain CFT073 in vivo. The new virulence factor which we designated TIR-containing protein C (TcpC) blocks Toll-like receptor signaling and the NLRP3 inflammasome signaling cascade by interacting with key components of both pattern recognition receptor systems (Cirl et al., 2008; Waldhuber et al., 2016). We infected wild type and knock-out mice with wildtype CFT073 and a mutant CFT073 strain lacking tcpC. This protocol describes how the mice were infected, how CFT073 was prepared and how the infection was monitored. The protocol was derived from our previously published work and allowed us to demonstrate that TcpC is a powerful virulence factor by increasing the bacterial burden of CFT073 in the urine and kidneys. Moreover, TcpC was responsible for the development of kidney abscesses since infection of mice with wildtype but not tcpC-deficient CFT073 mutants caused this complication.
Keywords: Uropathogenic Escherichia coliBackground
Urinary tract infections (UTIs) are some of the most common bacterial infections worldwide (Dielubanza and Schaeffer, 2011) and are predominantly caused by uropathogenic Escherichia coli (UPEC) (Zhang and Foxman, 2003). There is a high rate of recurrent infections (Dielubanza and Schaeffer, 2011) and also an increase in the emergence of antibiotic resistant E. coli strains (Eurosurveillance editorial, 2015). Therefore the understanding of host and bacterial factors in the pathophysiology of urinary tract infections is of high relevance in order to develop new therapeutic agents.
The murine UTI model system is the primarily used animal model system to study the pathogenicity of UPEC isolates and bacterium-host interactions and to identify underlying molecular mechanism. Besides the murine UTI model system, other animal model systems like porcine, avian, zebra fish and nematodes exist, which have been demonstrated to be useful for investigating UTIs. However, these models are associated with one or several limitations and disadvantages such as no possibility for genetic modification, the lack of a vertebrate-like immune system and/or urinary tract system or high costs. In addition to animal model systems cell culture based systems with primary immune cells or immortalized urinary tract tissue-derived cells are available. In vitro culture methods can be used to analyze UPEC interactions with host cells but they, of course, cannot reflect the complexity of the host environment involving a number of different cell types, tissue architecture and host defense mechanisms. Mice have much in common with humans including conserved immunological factors and a similar urinary tract system. Further, the availability of a variety of genetically distinct mouse strains to assess the impact of the genetic background makes the murine mouse model very accessible to study host-pathogen interactions in order to develop therapeutic agents.
Materials and Reagents
- Microcentrifuge tubes (1.5 ml for urine collection and 2.0 ml for tissue homogenization)
- 1 ml syringes (BD, catalog number: 309628 )
- 27 G needles (BD, catalog number: 305136 )
- ELISA plates (Nunc MaxiSorp® flat-bottom 96 well plate) (Sigma-Aldrich, catalog number: M9410 )
- Unfrosted or frosted glass slides (Thermo Fisher Scientific, Fisher Scientific, catalog number: 10149870 )
- poly-L-lysine-coated glass slides (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: P4981 )
- Coverslips 60 x 24 mm
- Mice
Only female mice were used for the UTI infection model because the urethra in male mice is extremely difficult to catheterize due to its anatomy. The following strain of mice at an age of 8 to 16 weeks was used:
C57BL/6
The following knock-out mice were successfully used in the model to study the relevance of a number of host-defense genes but these mice are optional and not required to generate the model:
Tlr4−/−
Myd88−/−
Trif−/−
TrifLps2/Lps2
Il-1β−/−
Irf3−/−
Note: All knock-out mice were at least 8x backcrossed to C57BL/6 mice. All mice were bred and housed in specific pathogen free conditions. Mice were only used upon permission of the local animal experimental ethics committee. - Bacteria
- UPEC strain CFT073, provided by ATCC (ATCC, catalog number: 700928 )
- tcpC-deficient CFT073 tcpC::kan, generated in the lab using the method described by Datsenko/Wanner (Datsenko and Wanner, 2000).
- Complemented mutant strain CFT073 tcpC::kan+pTcpC, the plasmid was generated as described (Cirl et al., 2008).
- UPEC strain CFT073, provided by ATCC (ATCC, catalog number: 700928 )
- Ice
- Dry ice
- Ethanol (70%)
- Isoflurane (Abbott Laboratories, Forene®, catalog number: 506949 )
- Ketamine (Ketaminol®, catalog number: 511519 )
- Xylazine (Rompun®, catalog number: 0 22545 )
- 0.9% NaCl
- Phosphate-buffered saline (PBS) (Sigma-Aldrich, catalog number: D8662 )
- Gentamicin
- Tissue-Tek® O.C.T. compound (SAKURA FINETEK, catalog number: 4583 )
- Tissue-Tek® Cryomold (SAKURA FINETEK, catalog number: 4565 )
- Isopentane (Sigma-Aldrich, catalog number: PHR1661 )
- Triton X-100 (Sigma-Aldrich, catalog number: X100 )
- Normal goat serum (Agilent Technologies, catalog number: X0907 )
- Rat monoclonal antibody NIMP-R14 (1: 200) (Abcam, catalog number: ab2557 )
- Antiserum to a synthetic peptide within the PapG adhesin (1:200), produced in the laboratory
- Goat anti-rat immunoglobulins (1:200), conjugated with Alexa 488 dye (A488; 495ext/519em nm) (Thermo Fisher Scientific, InvitrogenTM, catalog number: A-11006 )
- Goat anti-rabbit immunoglobulins (1:200), conjugated with Alexa 568 dye (A568; 578ext/603em nm) (Thermo Fisher Scientific, InvitrogenTM, catalog number: A-11011 )
- Anti-NLRP3 antibody (Santa Cruz Biotechnology, catalog number: sc-66846 )
- Anti-IL-1β antibody (Abcam, catalog number: ab9722 )
- Anti-ASC antibody (Abcam, catalog number: ab155970 )
- DAPI (Sigma-Aldrich, catalog number: D9542 )
- VECTASHIELD mounting medium (Vector Laboratories, catalog number: H-1000 )
- MIP-2 quantification kit (R&D Systems, catalog number: MM200 )
- Paraformaldehyde (PFA) (Sigma-Aldrich, catalog number: P6148 )
- Tryptic soy agar (TSA) (BD, DifcoTM, catalog number: 236950 )
- LB agar (BD, catalog number: 244520 )
- Columbia 5% blood agar (BD, catalog number: 221263 )
- Sucrose (Sigma-Aldrich, catalog number: 84100 )
- Ketamine/Xylazine solution (see Recipes)
- Media for cultivation of bacteria (see Recipes)
Tryptic soy agar plates, LB, Columbia 5% blood agar - Sucrose solution (see Recipes)
- 4% paraformaldehyde (see Recipes)
Equipment
- Burker chamber
- ELISA washer
- Drop glass jar for gas anesthesia
- Biosafety hood in biosafety level 2 facility
- Curved forceps, scissors
- Soft polyethylene catheter (0.61 mm outer diameter; Clay Adams) (BD, catalog number: 427401 )
- Stomacher 80 homogenizer (Seward medical, catalog number: 0080/000/EU )
- Cryostat sections were made with a Microtome blade C-35 (FEATHER Safety Razor, model: C-35)
- Fluorescence microscopy (Olympus, model: AX60 , equipped with filter sets [excitation/emission] 495ext/519em nm and 578ext/603em nm)
- AxioCam ERc 5s camera (Zeiss, model: AxioCam ERc 5s)
- Labsystems Multiskan PLUS reader (Analytical Instruments, Golden Valley, USA)
Procedure
- Infection model (direct instillation of bacteria into the bladder)
The infection protocol of mice was originally described by Hagberg et al. (1983) and was further modified by Cirl et al. (2008), Yadav et al. (2010) and Waldhuber et al. (2016).- To sample the urine the mouse will be placed to sit normally on the metal mesh of the cage. Upon lifting up the tale and hind legs, the mouse will hold on to the cage firmly with the front legs thus stretching the belly and exposing the underside. Mouse bladder is now emptied by gentle manual compression with index finger in the abdominal region (Figure 1) and urine is collected in sterile microcentrifuge tubes (kept on ice). Collected urine can be used to quantify colony-forming units (CFU) on TSA plates, for examination of inflammatory cells (e.g., neutrophils) or for cytokine analysis by ELISA. Alternatively, urine can be collected after anesthesia.
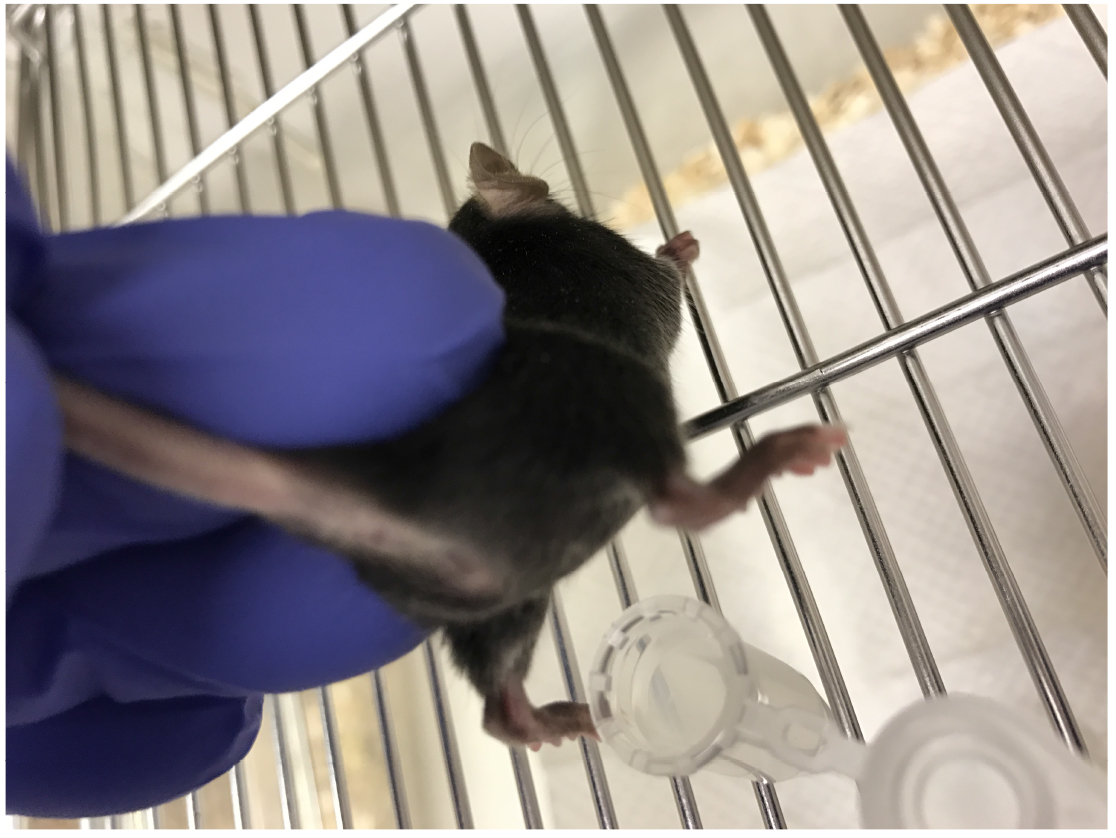
Figure 1. Urine collection in mice. Place the mouse on a cage metal mesh and hold tail to lift hind legs. Most mice will urinate naturally and urine can be collected in a sterile microcentrifuge tube. Sometimes, using index finger, gentle pressure on abdominal area can be applied to encourage mice to urinate. - The animals are anesthetized using either isoflurane in a drop glass jar or by intraperitoneal injection of ketamine-xylazine cocktail (Xylazin: 6-8 mg/kg body weight, Ketamin: 90-120 mg/kg body weight (normal body weight of 8-16 week old mice: 19-23 g) in 0.9% NaCl; diluted to a final volume of 100-200 µl/mouse for i.p. administration)
- If not done before anesthesia the bladder is now emptied by gentle compression of the abdomen. A drop of urine is caught directly at the urethral orifice with a calibrated loop (taking up 10 µl of fluid) and spread on blood agar plate to ensure that the mice are uninfected.
- Immediately thereafter, a soft polyethylene catheter (0.61 mm outer diameter; Clay Adams) adapted to a 0.4 by 20-mm gauge needle on a 1 ml tuberculin syringe (supplier BD) is transurethrally inserted into the bladder.
Note: By gently grabbing and pulling out the clitoral fold of the mouse, the urethra is straightened out and allows for fast and atraumatic catheterization. We use a forceps to pull the tissue prior to inserting the catheter with the mouse on the back (Figure 2).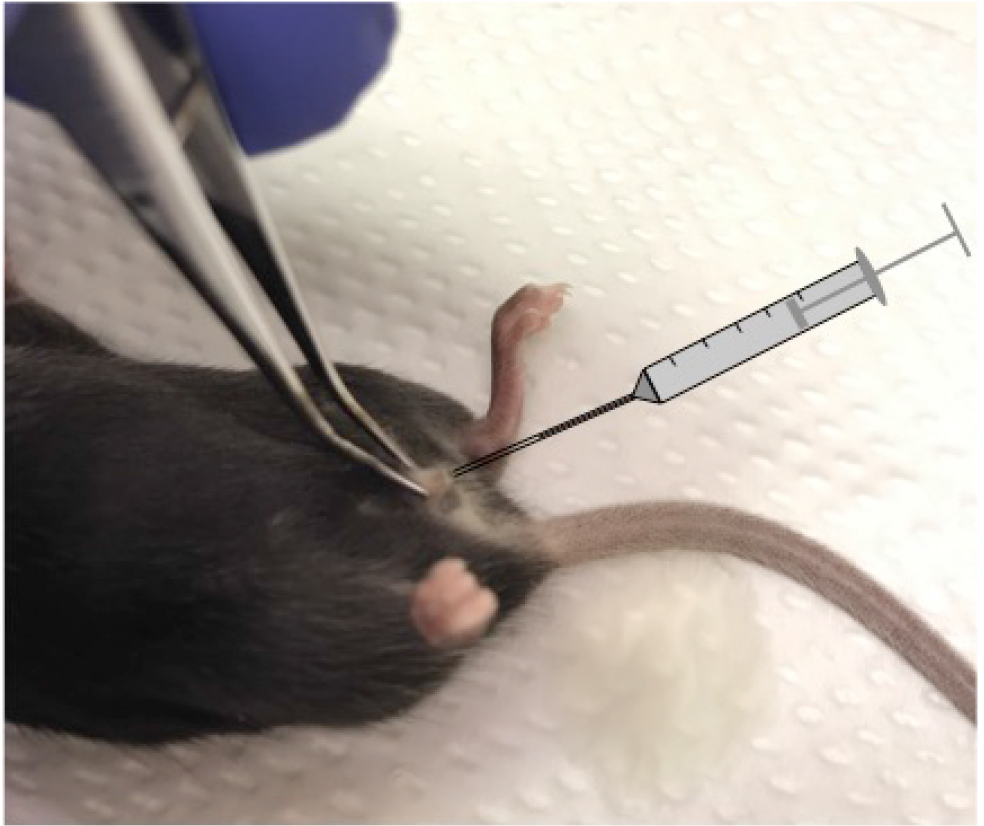
Figure 2. Murine urinary tract infection model. Image showing the position of the mouse during transurethral catheter insertion. - We used the high dose model in most experiments and instilled 0.1 ml of the bacterial suspension containing 2 x 108 or 1 x 109 CFU/ml. In a few experiments we lowered the bacterial amount to 2 x 105/mouse (low dose model) to study the effects of IL-1β deficiency on bacterial burden.
- After injection, the catheter is immediately withdrawn and no further manipulations are performed. Control mice were injected with sterile PBS.
- The model can then be used to study bladder and/or kidney infections (Figure 3).
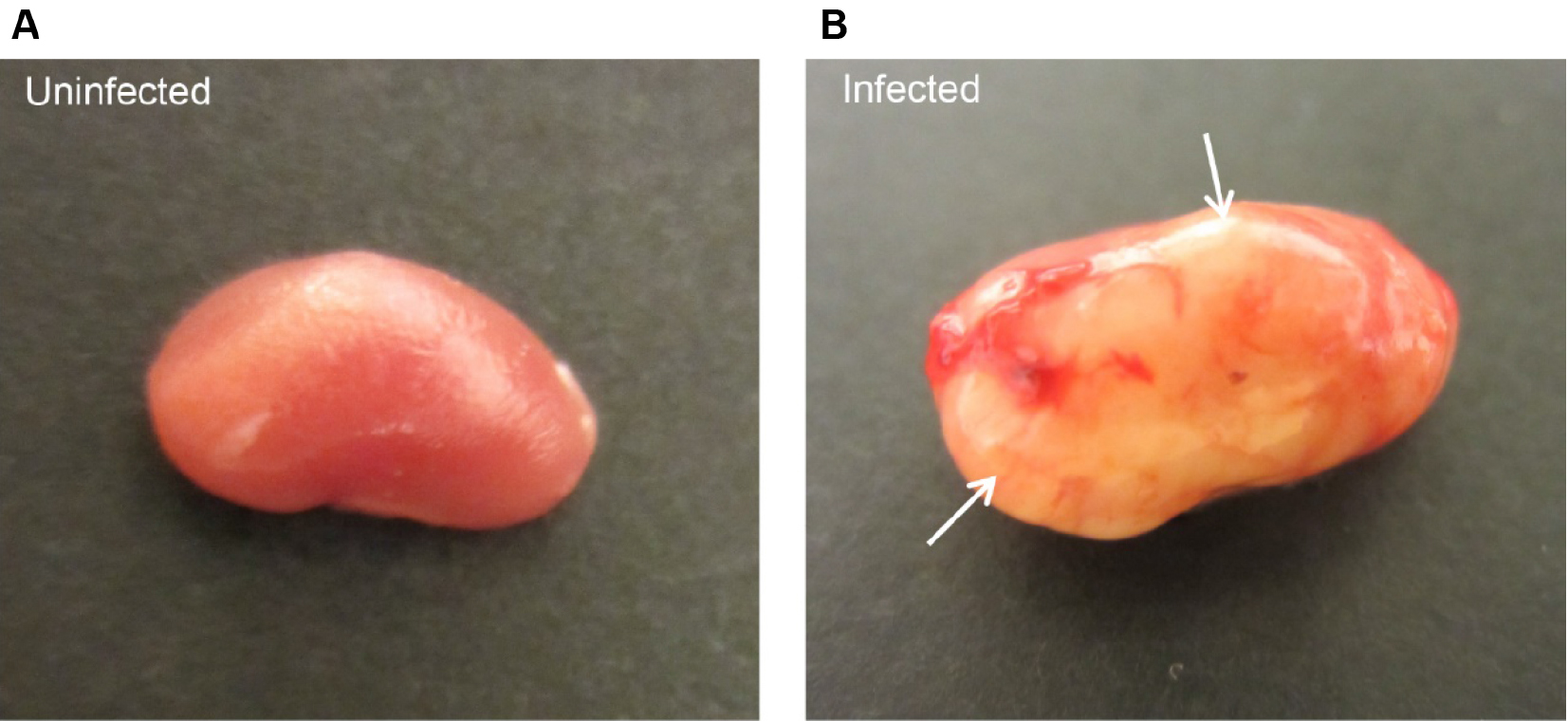
Figure 3. Ascending urinary tract infection in mice. Mice were infected with E. coli CFT073 by intravesical inoculation and sacrificed after 7 days. A. Uninfected control mice show healthy kidney with smooth and round contours, whereas kidney from infected mice displays severe abscess (arrows) formation (B).
- To sample the urine the mouse will be placed to sit normally on the metal mesh of the cage. Upon lifting up the tale and hind legs, the mouse will hold on to the cage firmly with the front legs thus stretching the belly and exposing the underside. Mouse bladder is now emptied by gentle manual compression with index finger in the abdominal region (Figure 1) and urine is collected in sterile microcentrifuge tubes (kept on ice). Collected urine can be used to quantify colony-forming units (CFU) on TSA plates, for examination of inflammatory cells (e.g., neutrophils) or for cytokine analysis by ELISA. Alternatively, urine can be collected after anesthesia.
- Monitoring infection
- The establishment and persistence of bacteria in the mouse urinary tracts is monitored by sequential urine cultures until sacrifice.
- Urine is collected into sterile tubes through gentle pressure on the mouse’s abdomen (5 h, 24 h and up to 7 days) and neutrophils are quantified by light microscopy using a Burker chamber.
- The urine samples and serial dilutions (starting dilution 1:100, then 1:1,000, 1:10,000 in PBS) are quantitatively cultured (100 μl) on tryptic soy agar (TSA) plates. LB or Columbia 5% blood agar plates are good alternatives. TSA plates do not contain antibiotics, since there is no antibiotic which would select between the experimental strain and any possible contamination. The experimental strain and possibly contaminating bacteria are differentiated by colony morphology. In addition, sham infected controls may also reveal possible contaminations, but these occur only rarely.
- After sacrifice by cervical dislocation, bladders and kidneys are removed under sterile conditions, placed into a plastic bag containing 5 ml PBS (pH 7.2) and homogenized in a Stomacher 80 homogenizer (100 rotations/min, 5 min; the instrument was cleaned with 70% alcohol between samples).
- Samples are placed on ice and analyzed in serial dilutions plated on TSA plates. The number of bacteria is expressed as the number of CFU per entire tissue.
- The number of intracellular bacteria is determined by killing extracellular bacteria with gentamicin (200 μg/ml) and subsequent washing and homogenization of the remaining organ (more information in Notes).
- Subsequently, blood agar and TSA plates are incubated at 37 °C overnight and visually scored for bacterial colonies.
- The establishment and persistence of bacteria in the mouse urinary tracts is monitored by sequential urine cultures until sacrifice.
- Post infection analysis
- Histology
- Kidneys and bladders are fixed in 1 ml of freshly prepared 4% PFA immediately after sacrifice and incubated overnight at 4 °C.
- Subsequently, the fixed tissues are incubated in 15% sucrose (4 °C/24 h) and washed in 25% ice-cold sucrose (4 °C/24 h). Tissues are then embedded in O.C.T. compound and frozen in isopentane at -40 °C and stored at -80 °C for further use. Cryostat sections (thickness 5 μm) are made with Microtome blade C-35, and mounted onto poly-L-lysine-coated glass slides and stained.
- The kidney sections from different groups of mice are double-stained for analysis by immunohistochemistry.
- Kidneys and bladders are fixed in 1 ml of freshly prepared 4% PFA immediately after sacrifice and incubated overnight at 4 °C.
- Immunohistochemistry
- Briefly, tissue sections are dried at 37 °C for 15 min, washed in PBS-0.2% Triton X-100 (pH 7.2) (2 x,10 min/RT) and incubated (30 min/RT) with PBS-0.2% Triton X-100 plus 5% goat normal decomplemented serum.
- Sections are subsequently incubated with a NIMP-R14 rat mAb (1:200) to detect neutrophils and a rabbit antiserum to the synthetic peptide CRPSAQSLEIKHGDL within the PapG adhesin (1:200) (produced in the lab) to detect UPECs for 2-3 h at RT. Negative controls consisted of only normal goat serum (1:200). Antibodies are diluted in PBS-0.2% Triton X-100 plus 5% goat normal serum.
- The kidney sections (thickness 5 μm) are washed in PBS (3 x, 5 min) by submersion in a washing chamber and incubated (1 h/RT) with secondary goat anti-rat immunoglobulins (1:200), conjugated with Alexa 488 (A488; 495ext/519em nm), and secondary goat anti-rabbit immunoglobulins (1:200), conjugated with Alexa 568 (A568; 578ext/603em nm) as fluorochrome.
- After washing in PBS (2 x, 5 min), specimens are counterstained (3 min/RT) with DAPI (0.05 µM, 1 ml) to stain cell nuclei.
- Sections are washed again in PBS (10 min) and mounted with VECTASHIELD mounting medium and kept in the dark (4 °C).
- Sections are analyzed by fluorescence microscopy (Carl Zeiss) with an AxioCam ERc 5s camera at 200x magnification.
- Alternative targets analyzed by immunohistochemistry included: E. coli, NLRP3, IL-1β, and ASC.
- Cytokine measurements
- Urine samples are collected at 0.6 h, 24 h and up to 7 days and stored at -20 °C.
- MIP-2 in urine is quantified by ELISA using the Mouse MIP-2 quantification kit according to the manufacturer's instructions. Urine is diluted five-fold in sample buffer.
- The ELISA plates are read at 450 nm in a Labsystems Multiskan PLUS reader.
- Histology
Data analysis
Parameters such as bacterial load in urine, urine cytokine content, number of polymorphonuclear leucocytes in urine were depicted as mean plus/minus standard deviation from at least 3 individual mice. The statistical difference of two groups or more were compared by Mann-Whitney rank sum test (Waldhuber et al., 2016), Student’s t-test (Yadav et al., 2010) or Fisher’s exact test (Cirl et al., 2008). Multiple groups were analyzed by 1-way ANOVA, post hoc Bonferroni’s multiple comparisons test (Cirl et al., 2008; Waldhuber et al., 2016).
Notes
Note: Intracellular detection of CFT073.
- Mice were infected transurethrally with CFT073.
- The bladder was removed micro-surgically at day 3 or 7 post infection.
- It was cut open by inserting a small scissor at the trigonum and it was further cut to the apex.
- If desired, adhering bacteria and residual urine was removed by flushing gently with 500 µl of PBS.
- The remaining cut open bladder was submersed in gentamicin solution (200 μg/ml, 15 min) to kill extracellular bacteria.
- Afterwards, the bladder was rinsed in PBS twice and the bladder was transferred into a microcentrifuge cup (2 ml, Eppendorf) containing PBS/Tween (0.3%, v/v) and a steel ball (4 mm stainless).
- The sample was stored on ice (critical to avoid overheating during homogenization) until homogenization was performed with a Retsch MM200 tissue homogenizer for 30 sec. with a frequency of 30/sec. Samples were placed back onto ice and analyzed in serial dilutions.
- The prepared material was plated on blood agar plates and individual colonies were counted.
Recipes
- Ketamine/Xylazine solution
Xylazine: 6-8 mg/kg body weight
Ketamine: 90-120 mg/kg body weight in 0.9% NaCl
Diluted to a final volume of 100-200 µl/mouse for i.p. administration - Media for cultivation of bacteria
- Tryptic soy agar plates
- Add 40 powder in 1 L distilled water
- Heat to boiling while stirring to dissolve powder
- Autoclave for 15 min at 121 °C to sterilize
- Allow to cool to about 50 °C. Pour into Petri dishes and allow to solidify (5 mm thickness)
- LB
- Add 35 g powder in 1 L distilled water
- Heat to boiling while stirring to dissolve powder
- Autoclave for 15 min at 121 °C to sterilize
- Allow to cool to about 50 °C. Pour into Petri dishes and allow to solidify (5 mm thickness)
- Columbia 5% blood agar
- These agar plates are available ready to use from BD (see Materials and Reagents)
- Sucrose solution
15% and 25% sucrose solution - 4% paraformaldehyde (500 ml)
- Take 400 ml of PBS in a glass beaker and keep on a stir plate inside a ventilated hood (paraformaldehyde is toxic)
- Heat the solution while stirring to approximately 60 °C
- Add 20 g of paraformaldehyde powder to the solution
- To dissolve paraformaldehyde powder, slowly add 1 N NaOH dropwise until the solution becomes clear. Cool down the solution and filter and adjust the volume to 500 ml with PBS
- Adjust the pH to 6.9 with diluted HCl
- Aliquot the solution and freeze at -20 °C and use within 2 months
Acknowledgments
TM was supported by the Deutsche Forschungsgemeinschaft (DFG) grant MI471/6-1. The protocol was initially developed by Hagberg et al. (1983) and modified in the publications of Cirl et al. (2008), Yadav et al. (2010) and Waldhuber et al. (2016). Authors declare that there are no conflicts of interest or competing interests that may impact the design and implementation of their protocol.
References
- Cirl, C., Wieser, A., Yadav, M., Duerr, S., Schubert, S., Fischer, H., Stappert, D., Wantia, N., Rodriguez, N., Wagner, H., Svanborg, C. and Miethke, T. (2008). Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins. Nat Med 14(4): 399-406.
- Datsenko, K. A. and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97(12): 6640-6645.
- Dielubanza, E. J. and Schaeffer, A. J. (2011). Urinary tract infections in women. Med Clin North Am 95(1): 27-41.
- Eurosurveillance editorial, t. (2015). ECDC publishes 2014 surveillance data on antimicrobial resistance and antimicrobial consumption in Europe. Euro surveillance 20(46).
- Hagberg, L., Engberg, I., Freter, R., Lam, J., Olling, S. and Svanborg Eden, C. (1983). Ascending, unobstructed urinary tract infection in mice caused by pyelonephritogenic Escherichia coli of human origin. Infect Immun 40(1): 273-283.
- Waldhuber, A., Puthia, M., Wieser, A., Cirl, C., Durr, S., Neumann-Pfeifer, S., Albrecht, S., Rommler, F., Muller, T., Zheng, Y., Schubert, S., Gross, O., Svanborg, C. and Miethke, T. (2016). Uropathogenic Escherichia coli strain CFT073 disrupts NLRP3 inflammasome activation. J Clin Invest 126(7): 2425-2436.
- Yadav, M., Zhang, J., Fischer, H., Huang, W., Lutay, N., Cirl, C., Lum, J., Miethke, T. and Svanborg, C. (2010). Inhibition of TIR domain signaling by TcpC: MyD88-dependent and independent effects on Escherichia coli virulence. PLoS Pathog 6(9): e1001120.
- Zhang, L. and Foxman, B. (2003). Molecular epidemiology of Escherichia coli mediated urinary tract infections. Front Biosci 8: e235-244.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Waldhuber, A., Puthia, M., Wieser, A., Svanborg, C. and Miethke, T. (2017). Analysis of the Virulence of Uropathogenic Escherichia coli Strain CFT073 in the Murine Urinary Tract. Bio-protocol 7(3): e2129. DOI: 10.21769/BioProtoc.2129.
Category
Microbiology > in vivo model > Bacterium
Immunology > Host defense > Murine
Cell Biology > Tissue analysis > Tissue isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link