- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Force Measurement on Mycoplasma mobile Gliding Using Optical Tweezers
Published: Vol 7, Iss 3, Feb 5, 2017 DOI: 10.21769/BioProtoc.2127 Views: 8321
Reviewed by: Sofiane El-Kirat-ChatelAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Analysis of Gram-negative Bacteria Peptidoglycan by Ultra-performance Liquid Chromatography
Laura Alvarez [...] Felipe Cava
Oct 5, 2020 5450 Views

Assessing Swarming of Aerobic Bacteria from Human Fecal Matter
Arjun S. Byju [...] Sridhar Mani
May 5, 2021 3908 Views

High-speed Atomic Force Microscopy Observation of Internal Structure Movements in Living Mycoplasma
Kohei Kobayashi [...] Makoto Miyata
Mar 5, 2022 3006 Views
Abstract
Dozens of Mycoplasma species, belonging to class Mollicutes form a protrusion at a pole as an organelle. They bind to solid surfaces through the organelle and glide in the direction by a unique mechanism including repeated cycles of bind, pull, and release with sialylated oligosaccharides on host animal cells. The mechanical characters are critical information to understand this unique mechanism involved in their infectious process. In this protocol, we describe a method to measure the force generated by Mycoplasma mobile, the fastest gliding species in Mycoplasma. This protocol should be useful for the studies of many kinds of gliding microorganisms.
Keywords: MycoplasmaBackground
Surface motility systems are spread over many bacterial species and they are not well elucidated compared to bacterial flagella and eukaryotic motor proteins (Jarrell and McBride, 2008), although potentially they can give us critical information to understand the survival strategy of bacteria. To elucidate a motility mechanism, we need information about the structure of machinery, the flow of energy, and the mechanical characters including speed and force. Optical tweezers are a special method used for micromanipulations or force measurements in the piconewton range under microscopy, by which an object with a diffractive index different from the medium is trapped at the center of focused laser beam (Ashkin et al., 1986). This method has greatly contributed to clarifying the features of motility systems including myosin, dynein, and kinesin, and now an established method in the field of biophysics. Here, we provide a protocol on how to measure force generated by surface moving microorganisms, based on our studies (Miyata et al., 2002; Tanaka et al., 2016) for gliding mechanism of M. mobile the fastest gliding species in class Mollicutes. This is the first protocol for force measurement made using optical tweezers in bio-protocol.
Materials and Reagents
- 18 x 18 mm Square microscope cover slip (Matsunami Glass, catalog number: C218181 )
- 22 x 40 mm Square microscope cover slip (Matsunami Glass, catalog number: C022401 )
- Double-sided tape (NICHIBAN, NICETACKTM, catalog number: NW-5 )
- Mending tape (3M, Scotch®, catalog number: 810-3-15 )
- Filter paper ϕ70 (ADVANTEC, catalog number: 00021070 )
- 1.5 ml microtube
- M. mobile 163K strain (ATCC, catalog number: 43663 )
- Polystyrene beads 1.0 µm in diameter (Polybead® Carboxylate Microspheres 1.00 µm) (Polysciences, catalog number: 08226-15 )
- PolyLink Protein Coupling Kit for COOH Microspheres (Polysciences, catalog number: 24350-1 )
- PolyLink coupling buffer
- PolyLink EDAC
- Avidin from egg white (Sigma-Aldrich, catalog number: A9275 )
- PBS with 20 mM glucose (PBS/G)
- PBS with 40 mM glucose
- Sulfo-NHS-LC-LC-biotin (Ez-LinkTM Sulfo-NHS-LC-LC-biotin) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 21338 )
- Glucose
- Nail polish
- Heart infusion broth (BD, catalog number: 238400 )
- Yeast extract (BD, catalog number: 212750 )
- 10 N NaOH
- Horse serum (Thermo Fisher Scientific, GibcoTM, catalog number: 16050122 )
- Amphotericin B (Sigma-Aldrich, catalog number: A2942 )
- Ampicillin Na (Nacalai Tesque, catalog number: 02739-32 )
- Sodium phosphate (pH 7.3)
- NaCl
- Aluotto medium (see Recipes)
- Phosphate-buffered saline (PBS) (see Recipes)
Equipment
- Centrifuge (Sigma Laborzentrifugen, model: Sigma 1-14 )
- 25 cm2 tissue culture flask (AS ONE, catalog number: 2-8589-01 )
- 25 °C incubator (Tokyo Rikakikai, model: LTI-400E )
- Sonicator (Emerson Industrial Automation, BRANSON, model: 2510J-MT )
- Optical microscope (Olympus, model: IX71 )
- Optical tweezers system (Note 1)
- High speed charge-coupled device (DigiMo, model: LRH2500XE-1 )
- Power meter (Laser Power/Energy Meter) (Coherent, model: FieldMaxII-TOP, catalog number: 1098580 )
- Autoclave
Software
- ImageJ (http://rsbweb.nih.gov/ij/index.html)
- IGOR Pro 6.33J (WaveMetrics, Portland, OR)
Procedure
- Coating polystyrene beads with avidin
- Warm polystyrene beads, coupling buffer and PBS to room temperature (RT).
- Centrifuge 60 µl polystyrene bead suspension at 2,000 x g for 6 min at RT.
- Discard supernatant, resuspend the pellet in 400 µl of coupling buffer and centrifuge it at 2,000 x g for 6 min at RT.
- Discard supernatant, resuspend the pellet in 160 µl of coupling buffer.
- Just before use, dissolve 11.7 mg of EDAC in 50 µl of coupling buffer, add 20 µl of this solution to the suspension and incubate for 5 min at RT.
- Dissolve 0.4 mg of avidin in 10 µl of coupling buffer to be 0.13 mM as the final concentration, add the avidin solution to the suspension and incubate it with gently end-over-end mixing for 3 h at RT.
- Centrifuge the suspension at 2,000 x g for 6 min at RT, discard the supernatant and resuspend the pellet in 450 µl of PBS. Repeat this step five times.
- Transfer the suspension to new microtube and storage it at 4 °C until use (it can be stored for 6 months).
- Warm polystyrene beads, coupling buffer and PBS to room temperature (RT).
- Biotinylation of M. mobile cell surface
- Inoculate 1 ml of frozen stock of M. mobile into 10 ml of Aluotto medium in a 25 cm2 tissue culture flask and statically cultivate it at 25 °C to reach an optical density at 600 nm of 0.06-0.08.
- Collect 9 ml of cultured cells by centrifugation at 12,000 x g for 4 min at RT, discard supernatant, resuspend the cell pellet in 900 µl of PBS/G and centrifuge it at 12,000 x g for 4 min at RT (Note 2).
- Just before use, prepare a 0.5 mM of Sulfo-NHS-LC-LC-biotin solution by dissolving 0.3 mg of Sulfo-NHS-LC-LC-biotin in 1 ml of PBS/G.
- Discard supernatant of step B2, resuspend the cell pellet (Note 2) in 300 µl of Sulfo-NHS-LC-LC-biotin solution and incubate the cell suspension for 15 min at RT (Hiratsuka et al., 2006).
- Centrifuge the suspension at 12,000 x g for 4 min at RT, discard supernatant, and resuspend the cell pellet (Note 2) in 300 µl of PBS/G. Repeat this step three times.
- Inoculate 1 ml of frozen stock of M. mobile into 10 ml of Aluotto medium in a 25 cm2 tissue culture flask and statically cultivate it at 25 °C to reach an optical density at 600 nm of 0.06-0.08.
- Force measurement by using optical tweezers
- Prepare a tunnel chamber (5-mm interior width, 18-mm length, 60-μm wall thickness) composed of two cover glasses from box, and double-sided tapes (Figure 1).
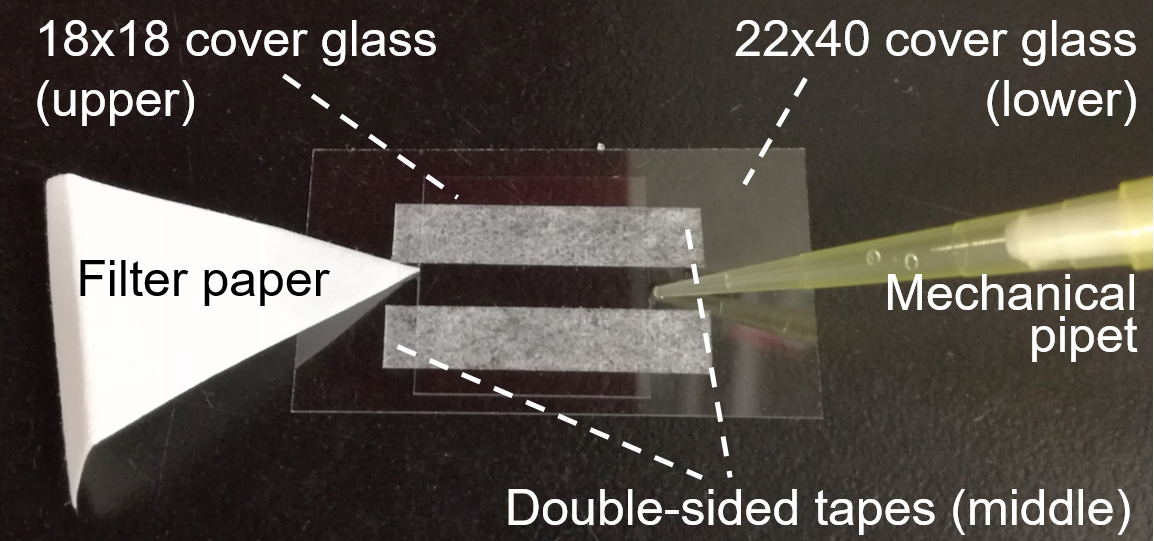
Figure 1. Tunnel chamber (step C1) - To coat the glass surface with glycoproteins, inject 20 µl of 10% horse serum in PBS/G into the tunnel chamber and incubate it for 15 min at RT (Note 3).
- Dilute the suspension of biotinylated cell with a 10 to 20-fold volume of PBS/G.
- Wash the tunnel chamber by flowing 40 µl of PBS/G, inject 20 µl of the diluted cell suspension into the tunnel chamber and incubate it for 15 min at RT.
- To remove floating cells, insert 20 µl of PBS/G into the tunnel chamber.
- Add the avidin-coated beads in PBS to the same volume of PBS containing 40 mM glucose, for adjusting glucose concentration to 20 mM.
- Dilute the bead suspension with PBS/G to get an appropriate bead density (10-20 beads per a 125.4 x 70.6 µm field of view) for the force measurements.
- Transfer 40 µl of bead suspension to a new microtube and sonicate it for 20 sec.
- Inject sonicated bead suspension into the tunnel chamber and seal both ends with nail polish.
- Set and fix the tunnel chamber by mending tapes onto the stage of inverted microscope equipped with optical tweezers (Figure 2).
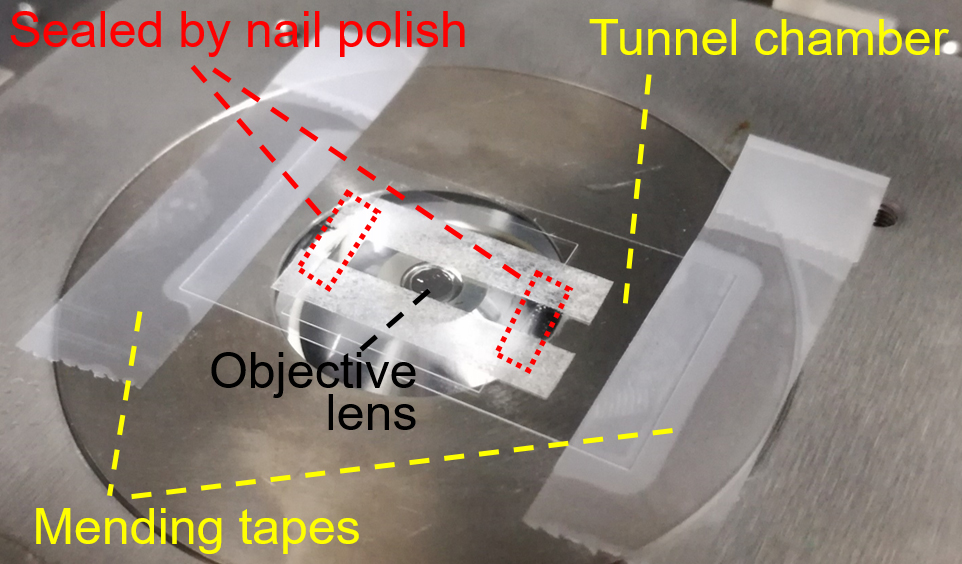
Figure 2. Setting tunnel chamber on microscope stage (step C10) - Trap a floating bead near the bottom of glass surface by focused laser beam, start to record the bead movements at 200 frames per second and immediately attached the bead to the back end of the gliding cell through the avidin-biotin interaction. The cell pulls the bead from trap center of optical tweezers and finally stalls (Video 1). The recording should be continued at least 30 sec after the start of stall (Note 4). After recording, turn off the laser beam.
- Remove the tunnel chamber and objective lens, turn on the laser beam to the same power as the step C11 and measure the applied laser power of the parallel light, which was achieved by the removal of objective lens, with using a power meter.
- Trace the bead movement from the trap center of optical tweezers and calculate the force from the distance between centers of the bead and the laser beam, by using ImageJ and IGOR Pro, as previously described (Tanaka et al., 2016).Video 1. Trapping of bead and attachment to gliding cell (step C11). Focused point of laser beam is indicated by a red circle, marked ‘Trap center’. At 5 sec, a flowing bead came from the right side and trapped at the beam. At 18 sec, a gliding cell was moved into the field by stage handling. At 27 sec, the bead was attached to the gliding cell. The cell pulled the trapped bead and stalled from 33 sec to the end.
- Prepare a tunnel chamber (5-mm interior width, 18-mm length, 60-μm wall thickness) composed of two cover glasses from box, and double-sided tapes (Figure 1).
Data analysis
The calculation method for bead positioning is described in Thompson et al. (2002).
Notes
- Construction of optical tweezers was described in Tanaka et al. (2016). Similar systems are also commercially available (Laser Optical tweezers – Mini type Basic model/without shutter, SIGMAKOKI, catalog number: LMS-M1064-2000. OTM200 Optical Tweezer Add-On, THORLABS, catalog number: OTM200).
- Suspension of cell pellet
More than 100 ups and downs with the pipette is recommended to separate cells completely. - Heat treatment of horse serum
For M. mobile growth and gliding, the horse serum should be inactivated by heat treatment at 56 °C for 30 min. - Available data size for ImageJ v1.43u is limited around 1 GB. The data size of movie depends on the recording time, the field size, and the frame rate. One example is 180 sec, 4 x 4 µm square, and 200 frames per second.
Recipes
- Aluotto medium
2.1% heart infusion broth
0.56% yeast extract
0.035% 10 N NaOH
Note: Dissolve the above three reagents as a mixture, autoclave, cool to lower than 37 °C, and add below three reagents in clean bench.
10% horse serum
0.025% amphotericin B
0.005% ampicillin Na - Phosphate-buffered saline (PBS)
75 mM sodium phosphate (pH 7.3)
68 mM NaCl
Sterilize by using filter or autoclave
Acknowledgments
This work was supported by Grants-in-Aid for Scientific Research on Innovative Area, ‘Harmonized Supramolecular Motility Machinery and Its Diversity’ (grant 24117002 to M. Miyata) and by a grant-in-aid for scientific research (B) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (grant 24390107 to M. Miyata).
References
- Ashkin, A., Dziedzic, J. M., Bjorkholm, J. E. and Chu, S. (1986). Observation of a single-beam gradient force optical trap for dielectric particles. Opt Lett 11(5): 288.
- Hiratsuka, Y., Miyata, M., Tada, T. and Uyeda, T. Q. (2006). A microrotary motor powered by bacteria. Proc Natl Acad Sci U S A 103(37): 13618-13623.
- Jarrell, K. F. and McBride, M. J. (2008). The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol 6(6): 466-476.
- Miyata, M., Ryu, W. S. and Berg, H. C. (2002). Force and velocity of mycoplasma mobile gliding. J Bacteriol 184(7): 1827-1831.
- Tanaka, A., Nakane, D., Mizutani, M., Nishizaka, T. and Miyata, M. (2016). Directed binding of gliding bacterium, Mycoplasma mobile, shown by detachment force and bond lifetime. MBio 7(3).
- Thompson, R. E., Larson, D. R. and Webb, W. W. (2002). Precise nanometer localization analysis for individual fluorescent probes. Biophys J 82(5): 2775-2783.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Mizutani, M. and Miyata, M. (2017). Force Measurement on Mycoplasma mobile Gliding Using Optical Tweezers. Bio-protocol 7(3): e2127. DOI: 10.21769/BioProtoc.2127.
Category
Microbiology > Microbial cell biology > Cell-based analysis
Microbiology > Microbial physiology > Membrane property
Cell Biology > Cell movement > Cell motility
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











