- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Protein Expression Protocol for an Adenylate Cyclase Anchored by a Vibrio Quorum Sensing Receptor
Published: Vol 7, Iss 2, Jan 20, 2017 DOI: 10.21769/BioProtoc.2112 Views: 9405
Reviewed by: Arsalan DaudiAgnès GroisillierTimo Lehti

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

From Llama to Nanobody: A Streamlined Workflow for the Generation of Functionalised VHHs
Lauren E.-A. Eyssen [...] Raymond J. Owens
Mar 20, 2024 6295 Views

Purification of Native Dentilisin Complex from Treponema denticola by Preparative Continuous Polyacrylamide Gel Electrophoresis and Functional Analysis by Gelatin Zymography
Pachiyappan Kamarajan [...] Yvonne L. Kapila
Apr 5, 2024 2117 Views

Thermus thermophilus CRISPR Cas6 Heterologous Expression and Purification
Junwei Wei [...] Yingjun Li
Jul 20, 2025 2196 Views
Abstract
The direct regulation of a mycobacterial adenylate cyclase (Rv1625c) via exchange of its membrane anchor by the quorum sensing receptor CqsS (Vibrio harveyi) has recently been reported (Beltz et al., 2016). This protocol describes the expression and membrane preparation for these chimeric proteins.
Keywords: Adenylate cyclase (AC)Background
Membrane-delimited mammalian adenylate cyclases (ACs) are class IIIa ACs. Regulation is indirectly via stimulatory (or inhibitory) Gα-proteins which are released intracellularly upon extracellular stimulation of G-protein-coupled-receptors (GPCR) by first messengers. ACs generate the universal second messenger cAMP using ATP as a substrate. The size of the two hexahelical membrane domains in vertebrate ACs by far exceeds the requirements for a simple membrane anchorage. Yet, regulatory features of this intrinsic membrane anchor/receptor domain are unknown. To investigate a potential function the canonical class IIIa AC Rv1625c from Mycobacteria was chosen which can be easily expressed in bacteria (Guo et al., 2001 and 2005) in contrast to mammalian AC isoforms. We replaced the hexahelical membrane anchor of the Rv1625c AC by the receptor domain of the hexahelical quorum sensing (QS) receptor from V. harveyi, CqsS, to examine whether we can confer a direct regulation of the AC by the QS-ligand ‘cholera autoinducer-1’, CAI-1. The design of the QS-receptor and the class IIIa membrane anchors are highly similar, i.e., minimal transmembrane α-helices and exceptionally short connecting loops.
We have demonstrated a direct regulation of a class IIIa AC by an extracellular signal. This considerably supports the hypothesis of a receptor function for the membrane anchor. Indeed, it raises the possibility that in addition to the well-established indirect GPCR-Gα-protein regulation of mammalian ACs a second, rather different set of signals directly impinge on this most important enzyme.
Materials and Reagents
- Expression
- FisherbrandTM syringe filter, 25 mm, 0.22 µm (Thermo Fisher Scientific, Fisher Scientific, catalog number: 09719A )
Note: This product has been discontinued. - Glycerol stock of Escherichia coli BL21 (DE3) (F-ompT hsdSB [rB-mB-] gal dcm [DE3]) transformed with plasmid pQE80L or pETDuet-3:
- pQE80L [QIAGEN, ΔXhoI, ΔNcoI]
Encodes a lacIq repression module; N-terminal RGS- His6-tag - pETDuet-3 (Figure 1)
MCS1 of pETDuet-1 [Novagen] is converted to MCS of pQE30 [QIAGEN]
MCS1: N-terminal RGS-His6-tag, MCS2: C-terminal S-tag
Note: Expression of both plasmids is controlled by an IPTG-inducible T5 (pQE80L) or T7 (pETDuet-3) promoter. pQE80L has only one MCS. If you want to co-express a second protein from the same plasmid, pETDuet expression vectors are suitable. - pQE80L [QIAGEN, ΔXhoI, ΔNcoI]
- Ampicillin sodium salt (Carl Roth, catalog number: K029 )
- IPTG (isopropyl β-D-thiogalactopyranoside) (AppliChem, catalog number: A1008 )
- LB broth (Lennox) (Carl Roth, catalog number: X964 )
- Ampicillin solution (see Recipes)
- IPTG solution (see Recipes)
- LB-medium (see Recipes)
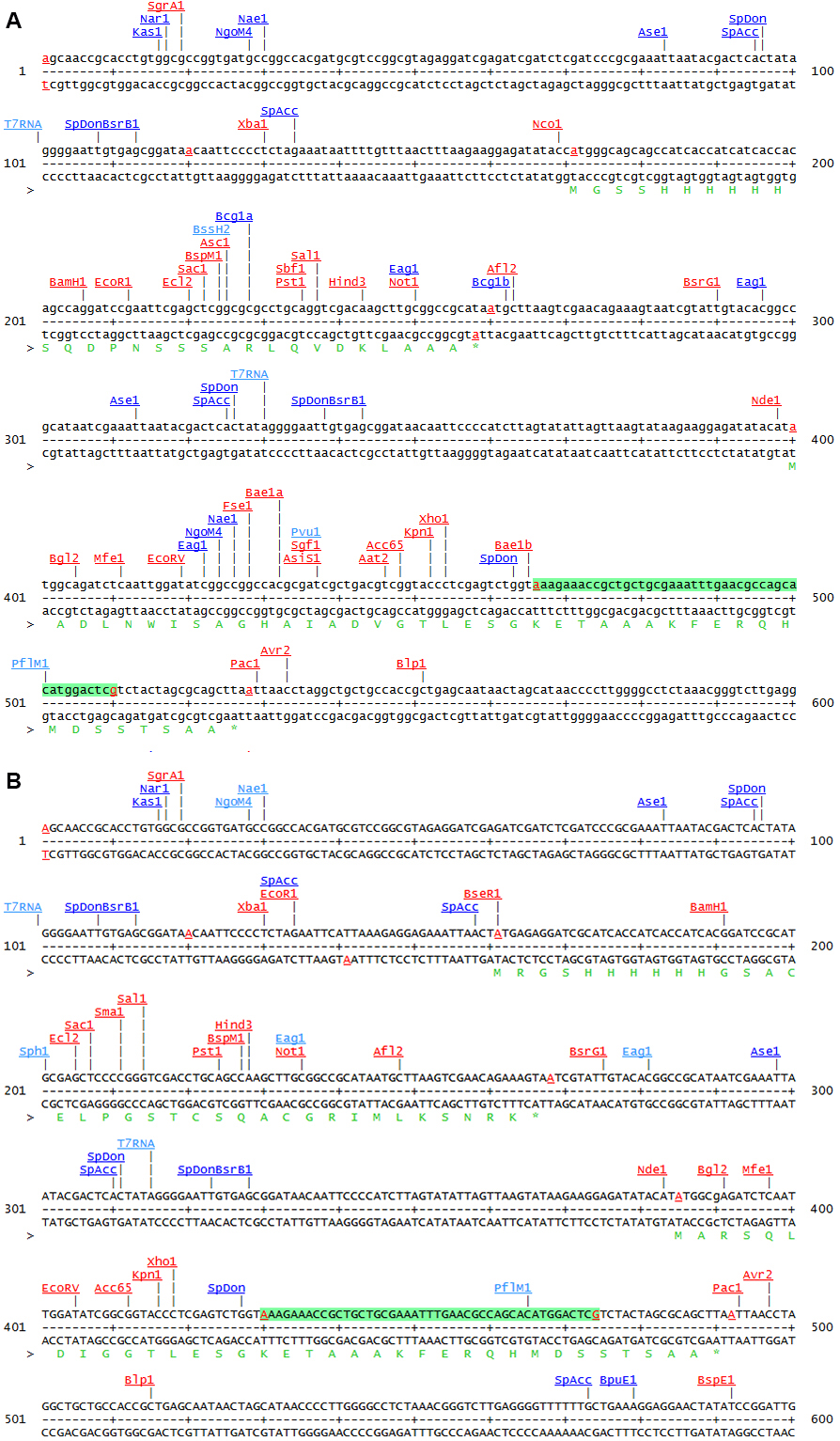
Figure 1. Comparison of the two MCS of pETDuet-1 (A) and pETDuet-3 (B)
Note: This sequence map is generated using the program DNA5 (https://pga.mgh.harvard.edu/web_apps/web_map/start).
- FisherbrandTM syringe filter, 25 mm, 0.22 µm (Thermo Fisher Scientific, Fisher Scientific, catalog number: 09719A )
- Cell harvesting
- Culture from expression
- Tris (AppliChem, catalog number: A1086 )
- HCl (37%)
- EDTA (Sigma-Aldrich, catalog number: E5134 )
- Wash-buffer (see Recipes)
- Membrane preparation
- 1.5 ml Eppendorf tube
- Frozen cell pellet from expression
- Thioglycerol (Sigma-Aldrich, catalog number: M1753 )
- Sodium chloride, NaCl (EMD Millipore, catalog number: 106404 )
- cOmpleteTM EDTA-free protease inhibitor (Roche Diagnostics, catalog number: 05056489001 )
- Glycerol (EMD Millipore, catalog number: 104094 )
- Liquid N2
- Lysis-buffer (see Recipes)
- Membrane-buffer (see Recipes)
- Data analysis
- Tris (AppliChem, catalog number: A1086)
- HCl (37%)
- Sodium dodecyl sulfate, SDS (Sigma-Aldrich, catalog number: 71729 )
- β-mercaptoethanol (EMD Millipore, catalog number: 805740 )
- Glycerol (EMD Millipore, catalog number: 104094)
- Bromophenol blue (Sigma-Aldrich, catalog number: B8026 )
- RGSHis-antibody (QIAGEN, catalog number: 34650 )
- S-tag-antibody (EMD Millipore, catalog number: 71549 )
- Protein marker IV prestained (VWR, catalog number: 27-2110 )
- Protein marker I unstained (VWR, catalog number: PEQL27-1010 )
- ECL Plex goat-anti-mouse IgG-Cy3-antibody (GE Healthcare, catalog number: PA43010 )
- 4x SDS-loading dye (see Recipes)
Equipment
- Expression
- 1 L Erlenmeyer flask
- Eppendorf Biophotometer
- Cuvettes
- Shaker incubator (37 °C and 22 °C)
- Cell harvesting
- Centrifuges (supplier: Thermo Fisher Scientific)
- Sorvall RC5B Plus
Rotor: Kontron-Hermle A6.14 (Sorvall, catalog number: 202200 ) - Heraeus Megafuge 1.0R
Rotor: Heraeus Sepatech BS4402/A (Heraeus, catalog number: 3360 ) - 250 ml Sorvall metal rotor tubes (Sorvall, catalog number: 522 )
- 50 ml centrifuge tube (Greiner Bio One, catalog number: 227261 )
- Vortex Genie-2 (VWR, catalog number: 4445900 )
- Box with water-ice-mix
- Freezer (-80 °C)
- Membrane preparation
- Vortex Genie-2 (Scientific Industries, model: Vortex-Genie 2 )
- French® Pressure Cell Press (SLM Instruments, SLM Aminco®, model: FA-078-E1 )
Alternative supplier: Glen Mills Inc. (USA) or G.Heinemann Ultraschall- und Labortechnik (Germany) - Aminco® French Pressure Cell (SLM Instruments, SLM Aminco®, catalog number: FA-073 , serial number: 9110668)
Alternative supplier: Glen Mills Inc. (USA) or G.Heinemann Ultraschall- und Labortechnik (Germany) - Centrifuges
- Heraeus Megafuge 1.0R
- Rotor: Heraeus Sepatech BS4402/A (Heraeus, catalog number: 3360 )
- Beckmann L-60
- Rotor: Beckman Coulter, model: Type 50.2 Ti
- Box with water-ice-mix
- Polycarbonate ultracentrifuge-tubes
- 7 ml Dounce Tissue Grinder (WHEATON, catalog number: 357542 )
- Freezer (-80 °C)
- Data analysis
- Ettan DIGE Imager (GE Healthcare, catalog number: 63005642 or 29-0834-61 )
- Ettan DIGE Imager (GE Healthcare, catalog number: 63005642 or 29-0834-61 )
Software
- Program DNA5 (https://pga.mgh.harvard.edu/web_apps/web_map/start)
Procedure
- Expression
- Inoculate a flask containing 200 ml LB-medium and 200 µl ampicillin (final concentration: 100 µg/ml) with approximately 5 ml overnight culture with the desired construct (see Materials and Reagents A1. point) to an OD600 of 0.1.
- Incubate the culture under shaking (200 rpm) at 37 °C up to an OD600 of 0.2-0.3 (approx. 45-90 min).
- Lower temperature to 22 °C (shaker incubator).
- At an OD600 of 0.4-0.6 induce expression by 500 µM IPTG (100 µl 1 M IPTG/200 ml culture).
- Cell harvesting
- Harvest the cells at an OD600 of 2.0-2.8 (120-150 min after induction).
Collect cells at 3,200 x g for 10 min at 4 °C (Sorvall centrifuge). - Add 25 ml of wash-buffer (4 °C) to the pellet, suspend by vortexing and pellet at 4,300 x g for 30 min at 4 °C (Heraeus centrifuge).
- Discard supernatant and store cells at -80 °C or continue with the membrane preparation.
- Harvest the cells at an OD600 of 2.0-2.8 (120-150 min after induction).
- Membrane preparation
- Thaw frozen cells on ice and suspend in 25 ml of lysis-buffer (4 °C) by vortexing.
Notes: - The cell pellet should be completely dissolved to avoid clogging the outlet of the Aminco® French Pressure Cell.
- Add always the cOmpleteTM EDTA-free protease inhibitor tablet just before using the lysis-buffer.
- Lyse cells mechanically by French press (1,100 psi) twice (Figure 2).
Notes: - Aminco® French Pressure Cell should be kept pre-cooled at 4 °C.
- Open outlet of the Aminco® French Pressure Cell such that a flow drop by drop is visible.
- Make sure that samples are continuously cooled in ice-water.
- Centrifuge homogenate for 30 min, 4 °C, 4,300 x g (Heraeus centrifuge) and discard pellet (cell debris).
- Transfer supernatant to an ultracentrifuge-tube and pellet membranes at 100,000 x g, 4 °C for 60 min (Beckman L-60 centrifuge).
- Decant supernatant, take up membranes (pellet) in 1-2 ml membrane-buffer and gently suspend in a homogenizer (Dounce Tissue Grinder). Transfer the membrane preparation into a 1.5 ml Eppendorf tube.
Note: The amount of membrane-buffer to be used depends on the size of the pellet. Suspend a pellet of approximately 1 cm in diameter at the bottom of the centrifuge tube in 2 ml membrane-buffer. - Freeze membrane preparation in liquid N2 and store at -80 °C.

Figure 2. Short French press protocol. A. Equipment of the French Pressure Cell; B. Aminco French Pressure Cell (taken out of the fridge 4 °C); C. Attention! Before use: Grease the O-rings and back-up rings of the stamp with glycerol! Insert the stamp into the cell body so that the inner cell body surface is covered with glycerol as well! D. Place the cell body with the stamp upside down into the stand with the opening facing upwards. E. Place the flow valve into the hole. F. Fill in the cell solution (looks milky). G. Put the lid on top. H. Close the flow valve. I. Place the French Pressure Cell into the gadget and… (J) close the bracket. K. Start the French press by turning the hand gear on ‘high’ and (L) switch on the pump. M. When the French press set up 1,100 psi… (N) open carefully the flow valve such that… (O) a flow drop by drop is visible. P. Attention! Stop in time so you can easily open the bracket and the stamp does not hit the ground! Q. When you are done, flip the switch to ‘down’ and turn on the pump again. The hydraulic lift moves down and you can take out the French Pressure Cell. R. Your cells are lysed properly when you can see the scale through the tube. S. Put your sample back on ice for the next step.
- Thaw frozen cells on ice and suspend in 25 ml of lysis-buffer (4 °C) by vortexing.
Data analysis
The expression of the proteins is verified by SDS-PAGE (Laemmli, 1970) and Western blot. The isolated membranes are incubated in 4x SDS-loading dye at room temperature for at least 30 min prior to application to SDS-PAGE (do not boil sample). Dilute the sample (membrane preparation) in MilliQ H2O to get 2.5-5 µg of protein in a final volume of 15 µl. Add 5 µl 4x SDS-loading dye. Load 20 µl of the mixture into one slot of the SDS-PAGE. The membranes are incubated for 1 h with each antibody (first antibody at 4 °C, second antibody at room temperature). The first antibody is either the RGSHis- (Figures 3A and 3C) or the S-tag-antibody (Figure 3B). In both cases, the ECL Plex goat-anti-mouse IgG-Cy3-antibody is used as a secondary antibody (dilution 1:2,500). Western blot evaluation is carried out using an Ettan DIGE Imager.
Note: In contrast to soluble proteins, membrane proteins are NOT boiled (95 °C, 5-10 min).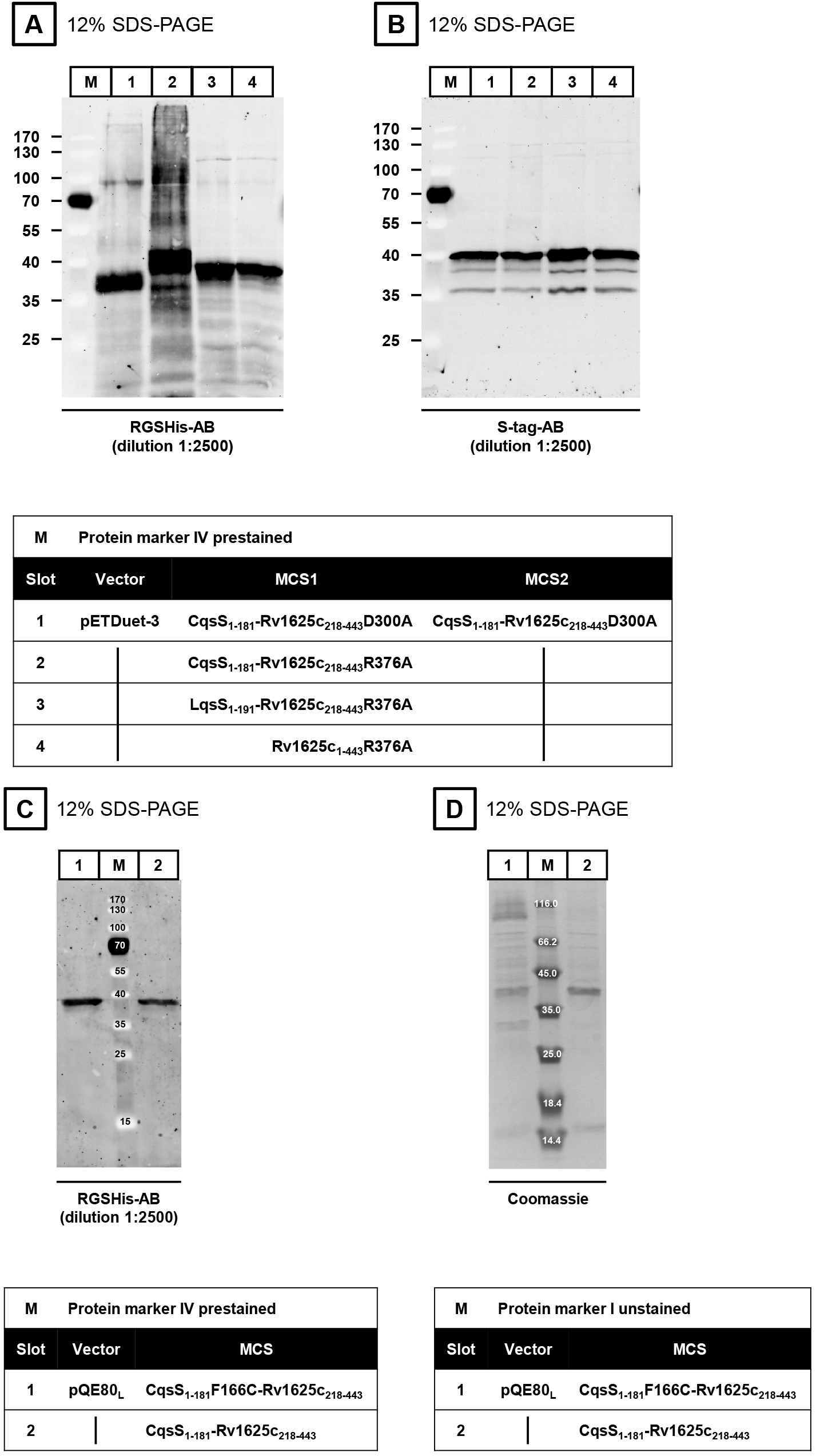
Figure 3. Western blots (A-C) and SDS-PAGE (D)
Recipes
- Ampicillin solution (100 mg/ml)
Dissolve 100 mg ampicillin in 1 ml MilliQ H2O (filter to sterilize) - 1 M IPTG solution
Dissolve 238.3 mg IPTG in 1 ml MilliQ H2O (filter to sterilize) - LB-medium
Dissolve 20 g LB in 1 L demineralized H2O (autoclave 20 min at 121 °C) - Wash-buffer
50 mM Tris/HCl (pH 8.0 at room temperature)
1 mM EDTA - Lysis-buffer
50 mM Tris/HCl (pH 8.0 at room temperature)
2 mM thioglycerol
50 mM NaCl
1 tablet cOmpleteTM EDTA-free protease inhibitor/50 ml lysis-buffer - Membrane-buffer
40 mM Tris/HCl (pH 8.0 at room temperature)
1.6 mM thioglycerol
20% glycerol (85%) - 4x SDS-loading dye
130 mM Tris/HCl (pH 6.8)
10% SDS
10% β-mercaptoethanol
20% glycerol (85%)
0.06% bromophenol blue
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 766; TP B08).
References
- Beltz, S., Bassler, J. and Schultz, J. E. (2016). Regulation by the quorum sensor from Vibrio indicates a receptor function for the membrane anchors of adenylate cyclases. Elife 5: e13098.
- Guo, Y. L., Kurz, U., Schultz, A., Linder, J. U., Dittrich, D., Keller, C., Ehlers, S., Sander, P. and Schultz, J. E. (2005). Interaction of Rv1625c, a mycobacterial class IIIa adenylyl cyclase, with a mammalian congener. Mol Microbiol 57(3): 667-677.
- Guo, Y. L., Seebacher, T., Kurz, U., Linder, J. U. and Schultz, J. E. (2001). Adenylyl cyclase Rv1625c of Mycobacterium tuberculosis: a progenitor of mammalian adenylyl cyclases. EMBO J 20(14): 3667-3675.
- Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227(5259): 680-685.
Article Information
Copyright
Beltz and Schultz. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Beltz, S. and Schultz, J. E. (2017). Protein Expression Protocol for an Adenylate Cyclase Anchored by a Vibrio Quorum Sensing Receptor. Bio-protocol 7(2): e2112. DOI: 10.21769/BioProtoc.2112.
- Beltz, S., Bassler, J. and Schultz, J. E. (2016). Regulation by the quorum sensor from Vibrio indicates a receptor function for the membrane anchors of adenylate cyclases. Elife 5: e13098.
Category
Microbiology > Microbial biochemistry > Protein > Isolation and purification
Microbiology > Microbial signaling > Quorum sensing
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









