- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Activity-based Pull-down of Proteolytic Standard and Immunoproteasome Subunits
(*contributed equally to this work) Published: Vol 6, Iss 24, Dec 20, 2016 DOI: 10.21769/BioProtoc.2073 Views: 9767
Reviewed by: Alka MehraMarielle CavroisAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Cluster FLISA—A Method to Compare Protein Expression Efficiency Between Cell Lines and Subunit Clustering of Proteins
Sabrina Brockmöller and Lara Maria Molitor
Nov 5, 2025 1209 Views

Detecting the Activation of Endogenous Small GTPases via Fluorescent Signals Utilizing a Split mNeonGreen: Small GTPase ActIvitY ANalyzing (SAIYAN) System
Miharu Maeda and Kota Saito
Jan 5, 2026 496 Views

Time-Lapse Into Immunofluorescence Imaging Using a Gridded Dish
Nick Lang [...] Andrew D. Stephens
Feb 20, 2026 50 Views
Abstract
Activity-based probes (ABP) are small organic molecules that irreversibly bind to the active center of a specific enzyme family and may be coupled to a fluorophore or an affinity tag (Li et al., 2013). Here, we describe a method to pull-down active catalytic standard and immunoproteasome subunits in cell lysates using the biotinylated, proteasome-specific ABP Biotin-Epoxomicin (Bio-EP). Covalent labeling of the active catalytic subunits with Bio-EP is followed by a pull-down using streptavidin-coated beads. After elution from the beads, enriched subunits may be detected via Western blot, tandem mass spectrometry (Li et al., 2013), or alternative techniques.
Keywords: Activity-based probeBackground
The proteasome is a barrel-shaped, multicatalytic enzyme complex that is present in the nucleus and cytoplasm of eukaryotic cells. It is essential for protein degradation, including processing of antigenic peptides for MHC I presentation, and regulates many cellular processes (Kammerl and Meiners, 2016). In cells of hematopoietic origin, the standard (constitutive) proteasome is often replaced by the immunoproteasome (Meiners et al., 2014), which differs in the incorporation of the three distinct catalytically active β-subunits (Figure 1).
To study the molecular function of single catalytic subunits and to modulate physiological processes, the development of subunit-specific proteasome inhibitors is indispensable. Large progress has recently been made in this area by de Bruin et al. (2014). Specific immunoproteasome inhibitors have proven as potential drug candidates for the treatment of inflammatory and autoimmune disease. The inhibition of the immunoproteasome subunit β5i may alter cytokine production by activated monocytes and lymphocytes. In a mouse model of rheumatoid arthritis, this reversed signs of the disease (Muchamuel et al., 2009).
The protocol was developed in the context of a study that investigated the effects of selective inhibition of β5i on the polarization of alveolar macrophages (Chen et al., 2016). The activity-based pull-down of catalytic standard and immunoproteasome subunits using the pan-reactive ABP Bio-EP allowed us to confirm the specific inhibition of β5i by the inhibitor ONX-0914, previously developed by ONYX Pharmaceuticals.
The use of ABPs with alternative specificity may allow for activity-based pull-down of other enzyme families in a similar experimental approach.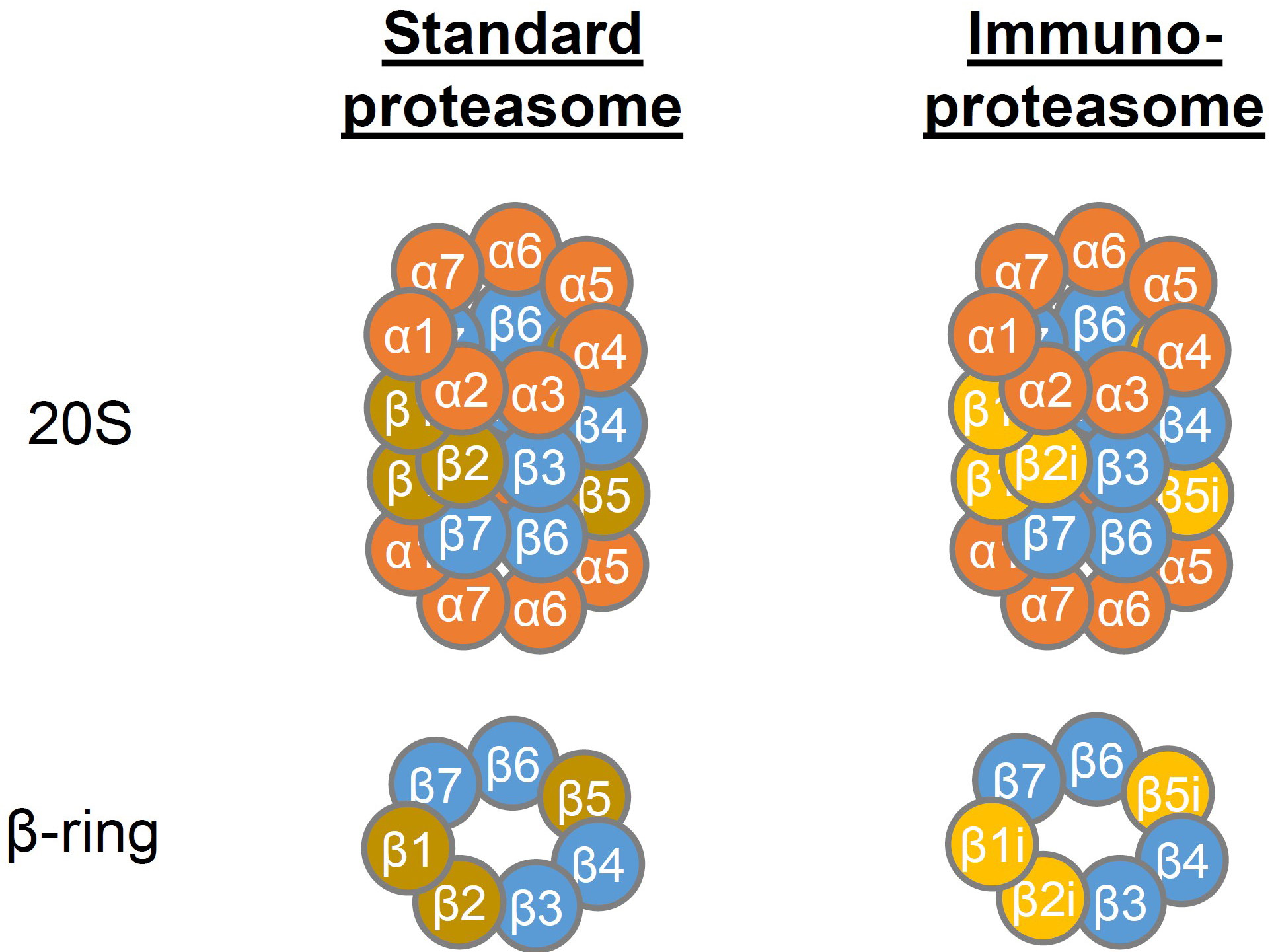
Figure 1. Structure of the 20S standard and immunoproteasome. The 20S core particle of the proteasome consists of two stacked inner rings containing the β-subunits 1-7 of which β1, β2 and β5 exhibit proteolytic activity. The β-rings are flanked by two α-rings containing the α-subunits 1-7. In the immunoproteasome β1, β2 and β5 are replaced by β1i, β2i and β5i, respectively, which leads to altered proteolytic activity.
Materials and Reagents
- 15 cm cell culture dishes
- Protein LoBind® tubes (1.7 ml tubes; OMNILAB-LABORZENTRUM, catalog number: 5409327 )
- 50 ml Falcon tube (Corning, Falcon®, catalog number: 352070 )
- Pipette tips
- Fine Dosage Syringe Omnifix-F (1 ml) (OMNILAB-LABORZENTRUM, catalog number: 5421736 )
- 15 ml Falcon tube (Corning, Falcon®, catalog number: 352096 )
- Immun-Blot® PVDF membrane (Bio-Rad Laboratories, catalog number: 162-0177 )
- PD MidiTrap G-25 (store at RT) (GE Healthcare, catalog number: 28-9180-08 )
- Filter 0.2 µm (Sartorius, catalog number 16534-K )
- Fuji X-ray films RX, 18 x 24 cm (Kisker Biotech, model: RX1824 )
- Whatman blotting paper (Laborbedarf Lammel, catalog number: 3030690 )
- Murine alveolar macrophage cell line MH-S (ATCC, catalog number: CRL-2019 )
- Liquid nitrogen
- BCA Protein Assay Kit (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 23225 )
- Biotin-Epoxomicin (Bio-EP; dissolved in DMSO, stable for at least 12 months at -20 °C; Hermen Overkleeft Lab, synthesis described in Florea et al., 2010)
- Dimethyl sulfoxide (DMSO) (Carl Roth, catalog number: A994.2 )
- HEPES (Molecular biology grade) (AppliChem, catalog number: A3724,1000 )
- Ultrapure water (e.g., MilliQ)
- Streptavidin beads (Strep-Tactin® Superflow® 50% suspension; store at 4 °C) (IBA, catalog number: 2-1206-010 )
- Methanol (AppliChem, catalog number: A3493.1000 )
- Roti®-Block blocking solution (store at RT) (Carl Roth, catalog number: A151.3 )
- Anti-β5 antibody (store at -20 °C) (Abcam, catalog number: 90867 )
- Anti-β5i antibody (store at -20 °C) (Abcam, catalog number: 3329 )
- Anti-Rabbit IgG, HRP-linked antibody (store at -20 °C) (New England Biolabs, catalog number: 7074S )
- Amersham ECL prime Western blotting detection reagent (store at 4 °C) (GE Healthcare, catalog number: RPN2232 )
- RPMI-1640 cell culture medium (Thermo Fisher Scientific, GibcoTM, catalog number 11875093 )
- Fetal bovine serum (FBS) (Biochrom, catalog number S 0615 )
- β-mercaptoethanol (Molecular biology grade) (AppliChem, catalog number A1108-100 )
- Penicillin-streptomycin (Thermo Fisher Scientific, GibcoTM, catalog number 15140122 )
- Tris (buffer grade) (AppliChem, catalog number: A1379,1000 )
- Magnesium chloride 6-hydrate (MgCl2.6H2O) (AppliChem, catalog number: A1036,0500 )
- Sodium chloride (NaCl) (AppliChem, catalog number: A2942,1000 )
- Ethylenediaminetetraacetic acid (EDTA) (AppliChem, catalog number: A2937,1000 )
- Sodium azide (NaN3) (AppliChem, catalog number: A1430,0100 )
- Dithiothreitol (DTT) (molecular biology grade) (AppliChem, catalog number: A2948,0025 )
- Adenosine triphosphate (ATP), disodium salt (10 g) (Roche Diagnostics, catalog number: 10127531001 )
- Glycerol (87%) (molecular biology grade) (AppliChem, catalog number: A3739,1000 )
- cOmpleteTM protease inhibitor cocktail (Roche Diagnostics, catalog number: 11697498001 )
- PhosSTOPTM (phosphatase inhibitor) (Roche Diagnostics, catalog number: 4906845001 )
- Sodium dodecyl sulfate (SDS) (pure) (AppliChem, catalog number: A1502,1000 )
- Bromophenol blue (AppliChem, catalog number: A2331,0025 )
- Hydrochloric acid (fuming) (37%) (Sigma-Aldrich, catalog number: 258148-2.5L )
- Rotiphorese® Gel 30 (37.5:1) (Carl Roth, catalog number: 3029.2 )
- Ammonium peroxodisulfate (APS) (AppliChem, catalog number: A0834,0250 )
- Tetramethylethylenediamine (TEMED) (AppliChem, catalog number: A1148,0100 )
- Tween 20 (Moleculare biology grade) (AppliChem, catalog number: A4974,1000 )
- Dulbecco’s phosphate-buffered saline (1x DPBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 14190-144 )
- Complete growth medium for MH-S cells (see Recipes)
- TSDG buffer (see Recipes)
- 500 µM Bio-EP in DMSO (see Recipes)
- 50 mM HEPES (pH 7.4) (see Recipes)
- 10% SDS (see Recipes)
- 6x Laemmli buffer (see Recipes)
- 4x SDS-PAGE resolving buffer (pH 8.8) (see Recipes)
- 4x SDS-PAGE stacking buffer (pH 6.8) (see Recipes)
- Stacking gel (see Recipes)
- Resolving gel (15% acrylamide) (see Recipes)
- PBST (0.1% Tween) (see Recipes)
Equipment
- Falcon tube centrifuge (Hettich Instruments, model: Rotina 420R )
- Table centrifuge (Hettich Instruments, model: MIKRO 200R )
- Water bath (LAUDA, model: Aqualine AL12 LCB 0725 )
- Thermomixer (Eppendorf, model: Thermomixer comfort )
- Forceps
- Pipettes
- Scissors
- Intelli-mixer rotator (ELMI, model: RM 2M )
- Western blot chambers (Bio-Rad Laboratories, model: Mini PROTEAN Tetra Cell )
- Power supply (PowerPacTM Basic Power supply) (Bio-Rad Laboratories, model: 1645050 )
- Western blot developer (Agfa-Gevaert, model: Curix60 )
- Fume hood
Procedure
- Preparation of cell material
- Culture MH-S cells in complete growth medium.
Note: This protocol was tested in the murine alveolar macrophage cell line MH-S purchased from ATCC (No. CRL-2019). Cells were passaged at 70-90% cell density in a 1:6 up to 1:12 ratio (splitting frequency approx. 3-5 days). 12 x 106 cells were seeded in 15 cm cell culture dishes the day before treatment, but numbers may vary if other cell lines are used. Further information about cell culture and treatment are stated in Chen et al. (2016). - Harvest cells and pellet at 800 x g for 5 min at RT, remove supernatant.
- Wash cells with 5 ml of PBS and pellet at 800 x g for 5 min at RT.
- Re-suspend cells gently in 400 µl of TSDG buffer.
Note: Lysis in TSDG buffer helps to sustain standard and immunoproteasome activity by stabilizing proteasome complexes. - Lyse cells with seven repeated cycles of freezing tubes in liquid nitrogen and thawing them in a water bath with water at room temperature until crystals disappear.
Note: The use of a floater facilitates the handling of several tubes in parallel. - Centrifuge at 14,000 x g for 20 min to remove cell debris and transfer the supernatant to a fresh tube.
- Determine protein concentration (for example with BCA protein assay, Bio-Rad).
- Store samples at -80 °C.
- Culture MH-S cells in complete growth medium.
- ABP labeling of the standard and immunoproteasome
- Treatment with Biotin-Epoxomicin (Bio-EP)
Note: Bio-EP is a biotinylated ABP that irreversibly binds to active catalytic subunits of both standard and the immunoproteasome.- Thaw cell lysate on ice and transfer volume equivalent to 500 µg of protein into a 1.7 ml tube.
Note: For each cell lysate, prepare two tubes. One tube serves as vehicle control and will be incubated with DMSO instead of Bio-EP. - Fill up to 500 µl with 50 mM HEPES (pH 7.4). The final protein concentration is 1 mg/ml.
- Add Bio-EP at a final concentration of 5 µM (for example 5 µl of a 500 µM stock), respectively, add the same amount of DMSO to the control tube.
- Incubate tubes for 120 min at 37 °C on a Thermomixer at 600 rpm.
Note: Meanwhile, equilibrate the columns for size exclusion chromatography (steps B2a-B2e) and prepare the beads for pull-down (steps C1a-C1d). - Fill up to 1 ml with 50 mM HEPES (pH 7.4). The final protein concentration is 0.5 mg/ml.
Note: A volume of 1 ml for SEC conforms manufacturer’s recommendation.
- Thaw cell lysate on ice and transfer volume equivalent to 500 µg of protein into a 1.7 ml tube.
- Size exclusion chromatography (SEC)
Note: SEC is performed to remove unbound ABP from the sample. Free ABP is retained in the column resin whereas large proteins like the standard and immunoproteasome pass through. Use a fresh PD MidiTrap G-25 column for each sample. Perform SEC also on vehicle control samples.- Remove the top cap from the column and pour off the storage solution.
- Use forceps to remove the filter on top of the column resin.
- Remove the bottom cap and insert the column into a 50 ml collection tube using the blue adapter delivered by the manufacturer.
- Equilibrate column 2 x with 5 ml of ultrapure water and discard the flow-through.
- Fill the column a third time with ultrapure water and spin down at 1,000 x g for 2 min in a centrifuge at RT.
Note: Don’t let the column run dry for a longer period. Cap the wet column with the bottom cap before centrifugation to pause equilibration until all samples are ready for application. Remove the cap and centrifuge the remaining liquid to resume with the protocol as soon as all samples are ready. - Discard the flow through and transfer the column with the adapter to a fresh 50 ml Falcon tube labeled with the samples name.
- Slowly pipette the sample dropwise in the middle of the packed bed and centrifuge at 1,000 x g for 2 min.
- Discard the column and store the 50 ml Falcon tube containing the eluate on ice.
- Remove the top cap from the column and pour off the storage solution.
- Treatment with Biotin-Epoxomicin (Bio-EP)
- Pull-down of ABP-labeled subunits
- Preparation of streptavidin beads
Note: Can be done during ABP incubation (step B1d).- Transfer 40 µl of the 50% streptavidin bead slurry (= 20 µl of beads) to a 1.7 ml tube.
Note: Vortex slurry before transfer. Use a 200 µl pipette and cut the end of the pipette tip with scissors, otherwise beads will plug the pipette tip. - Wash beads 3 x with 500 µl of PBS. Centrifuge between washing steps at 2,000 x g for 2 min and discard the supernatant.
Note: Use a narrow needled syringe and carefully aspirate the supernatant without removing beads. The diameter of the needle tip must be smaller than the bead diameter. - Re-suspend beads in 1 ml of 50 mM HEPES (pH 7.4) and transfer them into a 15 ml Falcon tube.
Note: Re-suspend the beads in 500 µl of HEPES buffer and transfer to a 15 ml Falcon tube. Repeat this to transfer the residual beads and hence prevent bead loss. Use a 1,000 µl pipette and cut the end of the pipette tip with scissors, otherwise beads will plug the pipette tip. - Store the beads on ice until needed.
- Transfer 40 µl of the 50% streptavidin bead slurry (= 20 µl of beads) to a 1.7 ml tube.
- Solubilization of the standard and immunoproteasome
Note: Solubilization is necessary to disrupt the complex structure and to allow separation of ABP-labeled from non-labeled subunits.- Transfer 200 µl (equals max. 100 µg protein) per eluate (from SEC) into a fresh 1.7 ml tube. Store the rest of the eluate at -20 °C.
- Add 20 µl of 10% SDS and boil at 95 °C for 8 min in a Thermomixer. Cool down to RT.
- Centrifuge tubes 1 min at 10,000 x g at RT to collect evaporated liquid from the lid of the tube.
- Transfer 200 µl (equals max. 100 µg protein) per eluate (from SEC) into a fresh 1.7 ml tube. Store the rest of the eluate at -20 °C.
- Pull-down
Note: Bio-EP labeled subunits are purified in this step using streptavidin beads.- Transfer sample to a 15 ml Falcon tube with pre-washed beads (from C1) and fill up to 10 ml with 50 mM HEPES (pH 7.4).
Note: SDS may interfere with the pull-down. Therefore, SDS is diluted to 0.02% in this step. - Incubate tubes for 2 h at 4 °C. Rotate tubes in a rotator during incubation (app. 20 rpm).
- Centrifuge tubes at 2,000 x g for 2 min (4 °C). Discard the supernatant.
- Re-suspend beads in 2 x 500 µl of PBS and transfer slurry into a 1.7 ml tube.
- Centrifuge tubes at 2,000 x g for 2 min (4 °C). Discard the supernatant.
- Wash beads two more times with 500 µl of PBS. Centrifuge at 2,000 x g for 2 min (4 °C).
- Re-suspend beads in 60 µl of 1x Laemmli buffer and boil for 15 min at 90 °C in a Thermomixer.
Note: Bio-EP labeled subunits bound to the beads are eluted in this step. - Centrifuge tubes at 2,000 x g for 2 min (4 °C). Transfer the supernatant into a fresh 1.7 ml tube using a syringe.
- Store samples at -20 °C.
- Transfer sample to a 15 ml Falcon tube with pre-washed beads (from C1) and fill up to 10 ml with 50 mM HEPES (pH 7.4).
- Preparation of streptavidin beads
- Western blot detection
Note: Alternative modes of detection or further processing steps may be performed. The protocol exemplifies the detection of immunosubunit β5i and standard proteasome subunit β5.- Separate proteins of the pull-down eluate in a 15% polyacrylamide gel. Load gel pocket with 25 µl of eluate and run for approximately 90 min at 120 V.
- Blot proteins onto a polyvinylidene difluoride membrane for 90 min at 250 mA.
Note: Soak membrane in MeOH before performing the electroblot. - Block the membrane for 60 min at RT in 1x Roti®-Block blocking solution.
- Incubate membrane with either:
a.1:2,000 anti-β5i (in Roti®-Block) overnight/over the weekend at 4 °C;
b.1:1,000 anti-β5 (in Roti®-Block) overnight/over the weekend at 4 °C. - Wash membrane three times with PBST for 10 min in a plastic box.
- Incubate with 1:40,000 anti-rabbit HRP-coupled secondary antibody (New England Biolabs, diluted in PBST) for 60 min at RT.
- Wash membrane 3 x with PBST for 10 min at RT.
- Cover the membrane with freshly prepared ECL substrate.
- Place membrane in a plastic foil and cover it with an X-ray film in a dark chamber. Develop film after several seconds to minutes.

Figure 2. Workflow activity-based pull-down. Active catalytic standard (brown) and immunoproteasome (yellow) subunits are covalently labeled with the ABP Bio-EP. After removal of unbound ABP via SEC the complex structure of the proteasome is solubilized in 1% SDS. A pull-down with streptavidin-coated agarose beads is performed to capture ABP-labeled subunits. Non-labeled proteins are removed in three consecutive steps of PBS washing, bound subunits are eluted via heating in 1x Laemmli.
- Separate proteins of the pull-down eluate in a 15% polyacrylamide gel. Load gel pocket with 25 µl of eluate and run for approximately 90 min at 120 V.
Data analysis
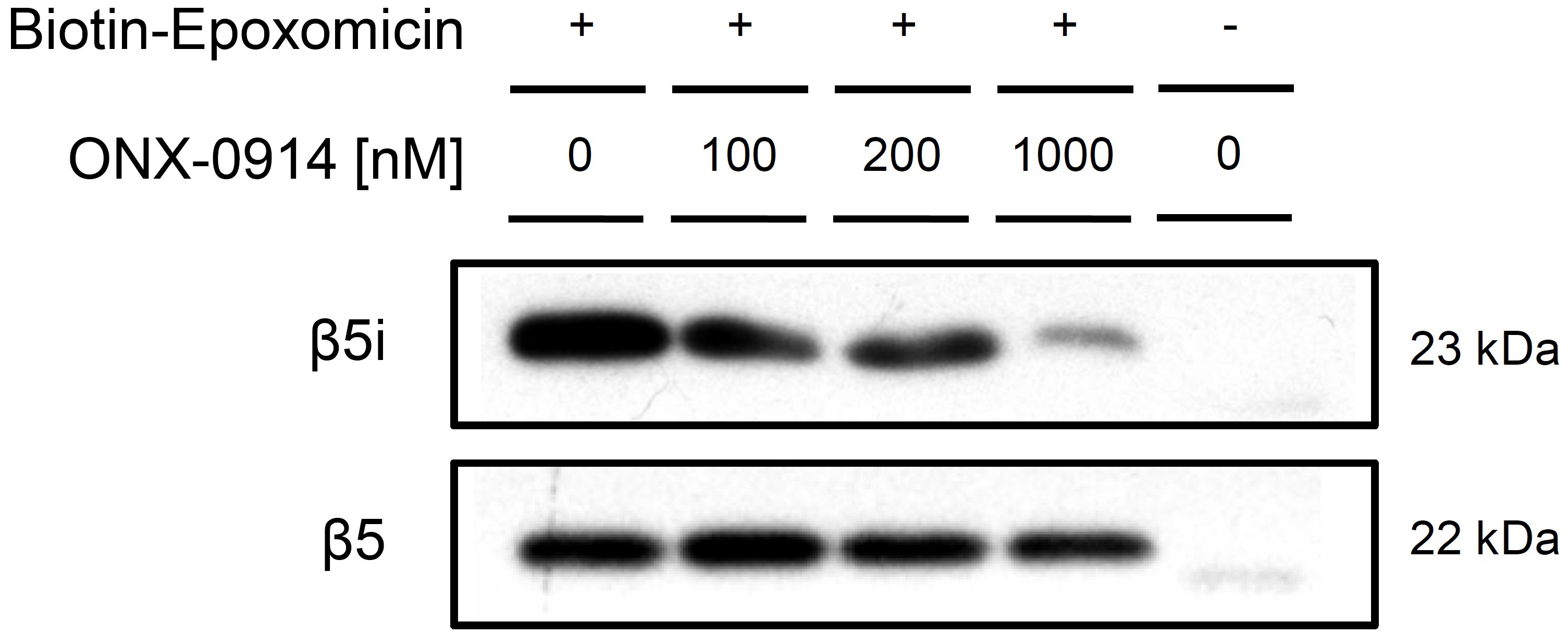
Figure 3. Western blot of activity-based pull-down with Bio-EP. Western blots show the amount of active β5i and β5 in ONX-0914 treated MH-S cell lysates and are representative for three independent experiments. With increasing concentration of the β5i-specific inhibitor, the amount of detected β5i in the pull-down eluate decreases whereas β5 remains unaffected by ONX-0914 treatment.
Recipes
- Complete growth medium for MH-S cells
500 ml RPMI-1640
10% fetal bovine serum (FBS)
50 μM β-mercaptoethanol
100 U/ml penicillin-streptomycin - TSDG buffer (10 ml)
1 ml 100 mM Tris stock (pH 7.5)
1.1 ml 10 mM MgCl2 stock
100 µl 1 M NaCl stock
20 µl 50 mM EDTA stock
100 µl 100 mM NaN3 stock
10 µl 1 M DTT stock
100 µl 200 mM ATP stock
1.15 ml 87% glycerol
Fill up to 10 ml with ultrapure water
Final concentrations:10 mM Tris/HCl, 1.1 mM MgCl2, 10 mM NaCl, 0.1 mM EDTA, 1 mM NaN3, 1 mM DTT, 2 mM ATP, 10% (v/v) glycerol
Adjust pH to 7.0
Aliquot and store at -20 °C
Add protease and phosphatase inhibitor (Roche) immediately before use (1 tablet for 50 ml lysis buffer; alternatively, a 25x stock can be prepared in ultrapure water and freshly added as needed, store stock at -20 °C) - 500 µM Bio-EP in DMSO
1 mg Bio-EP (MW = 724.95)
2.759 ml DMSO
Aliquot (30-50 µl suggested) and store at -20 °C - 50 mM HEPES (pH 7.4; 1 L)
11.915 g HEPES
Fill up to 1 L with ultrapure water
Adjust pH to 7.4 with HCl or NaOH, as necessary - 10% SDS (10 ml)
1 g SDS
Fill up to 10 ml with ultrapure water
Prepare under a fume hood, wear appropriate breath protection. - 6x Laemmli buffer (10 ml)
3 ml 1 M Tris HCl (pH 6.8)
1.5 ml glycerol
0.6 g SDS
0.5 g DTT
1 mg bromophenol blue
Fill up to 10 ml with ultrapure water
Filter (0.2 µm)
Aliquot and store at -20 °C - 4x SDS-PAGE resolving buffer (pH 8.8)
36.4 g Tris
Fill up to 150 ml with ultrapure water
Adjust pH to 8.8 with 37% HCl
Fill up to 200 ml with ultrapure water
Filter (0.2 µm)
0.48 g SDS
Storage: RT - 4x SDS-PAGE stacking buffer (pH 6.8)
6.05 g Tris
Fill up to 40 ml with ultrapure water
Adjust pH to 6.8 with 37% HCl
Fill up to 100 ml with ultrapure water
Filter (0.2 µm)
0.4 g SDS
Storage: RT - Stacking gel (4 ml)
1 ml 4x stacking buffer
480 µl 30% acrylamide
2.52 ml ultrapure H2O
50 µl 10% APS
12 µl TEMED - Resolving gel (15% acrylamide) (8 ml)
2 ml 4x resolving buffer
4 ml 30% acrylamide
2 ml ultrapure H2O
100 µl 10% APS
12 µl TEMED - PBST (0.1% Tween) (1 L)
999 ml 1x PBS
1 ml Tween 20
Acknowledgments
This protocol was used for the work previously published in Cell Death and Differentiation (Chen et al., 2016). The work was supported by intramural funding of the Helmholtz Zentrum München. The authors declare no conflict of interest.
References
- Chen, S., Kammerl, I. E., Vosyka, O., Baumann, T., Yu, Y., Wu, Y., Irmler, M., Overkleeft, H. S., Beckers, J., Eickelberg, O., Meiners, S. and Stoeger, T. (2016). Immunoproteasome dysfunction augments alternative polarization of alveolar macrophages. Cell Death Differ 23(6): 1026-1037.
- de Bruin, G., Huber, E. M., Xin, B. T., van Rooden, E. J., Al-Ayed, K., Kim, K. B., Kisselev, A. F., Driessen, C., van der Stelt, M., van der Marel, G. A., Groll, M. and Overkleeft, H. S. (2014). Structure-based design of β1i or β5i specific inhibitors of human immunoproteasomes. J Med Chem 57(14): 6197-6209.
- Florea, B. I., Verdoes, M., Li, N., van der Linden, W. A., Geurink, P. P., van den Elst, H., Hofmann, T., de Ru, A., van Veelen, P. A., Tanaka, K., Sasaki, K., Murata, S., den Dulk, H., Brouwer, J., Ossendorp, F. A., Kisselev, A. F. and Overkleeft, H. S. (2010). Activity-based profiling reveals reactivity of the murine thymoproteasome-specific subunit beta5t. Chem Biol 17(8): 795–801.
- Kammerl, I. E. and Meiners, S. (2016). Proteasome function shapes innate and adaptive immune responses. Am J Physiol Lung Cell Mol Physiol 311(2): L328-336.
- Li, N., Kuo, C. L., Paniagua, G., van den Elst, H., Verdoes, M., Willems, L. I., van der Linden, W. A., Ruben, M., van Genderen, E., Gubbens, J., van Wezel, G. P., Overkleeft, H. S. and Florea, B. I. (2013). Relative quantification of proteasome activity by activity-based protein profiling and LC-MS/MS. Nat Protoc 8(6): 1155-1168.
- Meiners, S., Keller, I. E., Semren, N. and Caniard, A. (2014). Regulation of the proteasome: evaluating the lung proteasome as a new therapeutic target. Antioxid Redox Signal 21(17): 2364-2382.
- Muchamuel, T., Basler, M., Aujay, M. A., Suzuki, E., Kalim, K. W., Lauer, C., Sylvain, C., Ring, E. R., Shields, J., Jiang, J., Shwonek, P., Parlati, F., Demo, S. D., Bennett, M. K., Kirk, C. J. and Groettrup, M. (2009). A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat Med 15(7): 781-787.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Baumann, T., Vosyka, O., Florea, B. I., Overkleeft, H. S., Meiners, S. and Kammerl, I. E. (2016). Activity-based Pull-down of Proteolytic Standard and Immunoproteasome Subunits. Bio-protocol 6(24): e2073. DOI: 10.21769/BioProtoc.2073.
Category
Biochemistry > Protein > Immunodetection
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link