- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Lentiviral shRNA Screen to Identify Epithelial Integrity Regulating Genes in MCF10A 3D Culture
Published: Vol 6, Iss 23, Dec 5, 2016 DOI: 10.21769/BioProtoc.2050 Views: 12142
Reviewed by: HongLok LungSteve JeanAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Automated Analysis of Cell Surface Ruffling: Ruffle Quantification Macro
Nicholas D. Condon [...] Adam A. Wall
Jan 20, 2020 5198 Views

In Vivo Retroviral Transduction of Cardiac Myofibroblasts Using Intramyocardial Injection Immediately Post-myocardial Infarction
Satsuki Ono [...] Michio Nakaya
Nov 20, 2025 1561 Views

A Highly Efficient siRNA Transfection Method in Primary Cultured Cortical Neurons
Xiaorong Wang [...] Zhaolong Zhang
Jan 20, 2026 296 Views
Abstract
MCF10A 3D culture system provides a reductionist model of glandular mammary epithelium which is widely used to study development of glandular architecture, the role of cell polarity and epithelial integrity in control of epithelial cell functions, and mechanisms of breast cancer. Here we describe how to use shRNA screening approach to identify critical cell pathways that couple epithelial structure to individual cell based responses such as cell cycle exit and apoptosis. These studies will help to interrogate genetic changes critical for early breast tumorigenesis. The protocol describes a library of lentiviral shRNA constructs designed to target epithelial integrity and a highly efficient method for lentiviral transduction of suspension MCF10A cultures. Furthermore, protocols are provided for setting up MCF10A 3D cultures in Matrigel for morphometric and cellular response studies via structured illumination and confocal microscopy analysis of immunostained 3D structures.
Keywords: 3D culturesBackground
All epithelial cells form highly organized tissue structures, which provide physical support and a structured scaffold for coordinated cell signaling. Such coordinated signaling across the epithelial structures is fundamental for epithelial biology; enabling dynamic joint actions of epithelial cells in regulation of organ size, shape, function and individual cell based responses (Roignot et al., 2013; Shamir and Ewald, 2014). Joint command of epithelial signaling also presents a powerful tumor suppressor mechanism by gatekeeping extrinsic and intrinsic mitogenic signals to quiescent epithelial tissues (Partanen et al., 2013; Rejon et al., 2016). However, very little is still known about genetic mechanisms coupling the status of epithelial structure with individual epithelial cell functions. MCF10A 3D Matrigel culture system is a well-established genetically tractable model of mammary epithelial architecture that is widely used to explore epithelial context-dependent cell functions (Debnath and Brugge, 2005). However, individual structures in MCF10A 3D cultures are not fully uniform in size or symmetry, which makes high-throughput screens with shRNA or cDNA reagents challenging in this system. Here, we describe protocols that expand the use of MCF10A 3D culture system from single gene studies to cell pathway level perturbation studies. The protocols for medium-throughput 3D screen using validated lentiviral shRNAs were originally used in a screen designed to identify genes with epithelial integrity-linked proliferation functions (Marques et al., [2016], screen outlined in Figure 1). However, these protocols are suitable for any reverse genetic MCF10A 3D culture study within a range of about 50 perturbed genes of interest.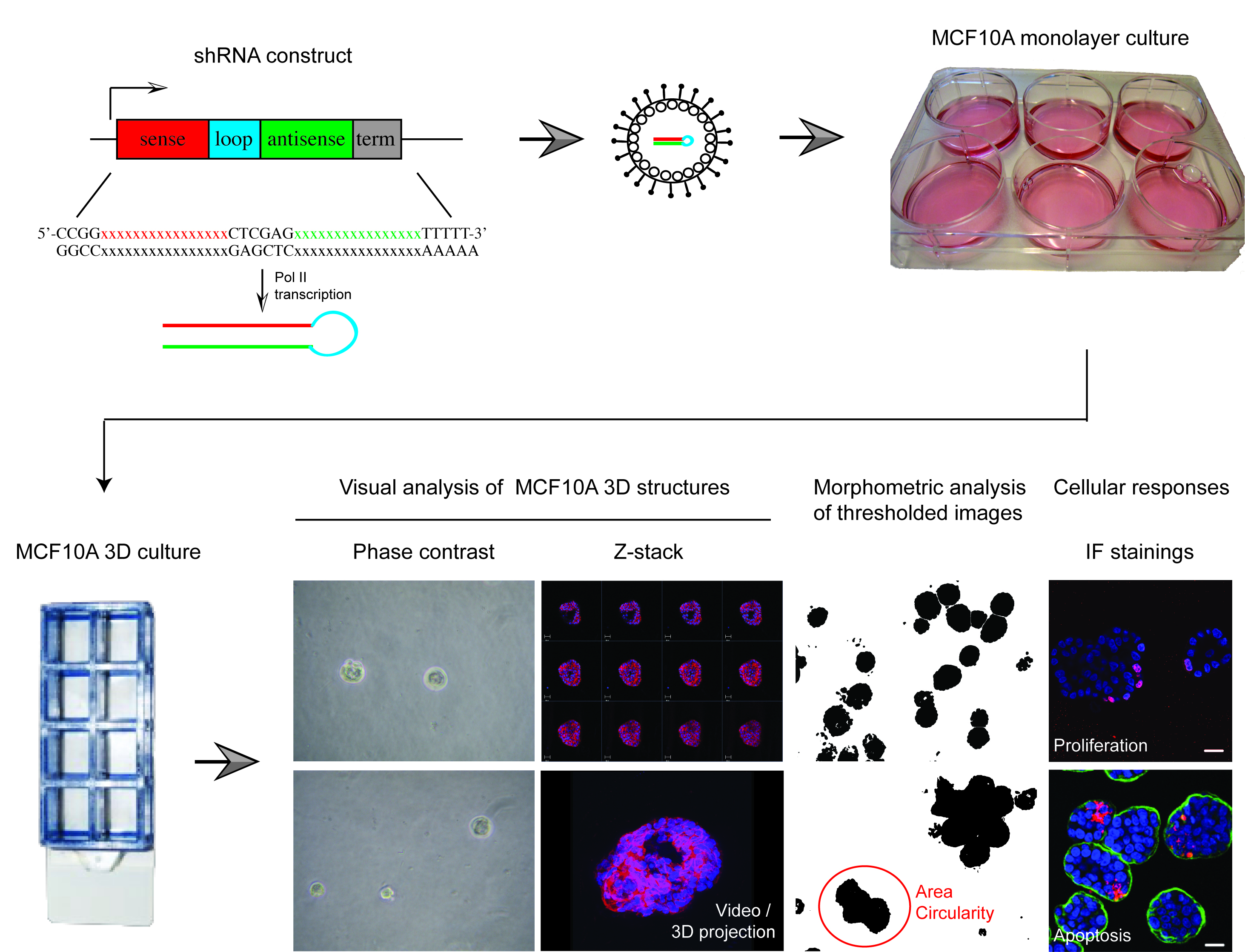
Figure 1. Overview of shRNA screen in MCF10A 3D culture designed to identify epithelial integrity regulating genes. The protocols described here were recently applied in a shRNA screen using 52 knockdown validated shRNAs, which were lentivirally transferred to MCF10A cells containing a switchable oncogenic form of Myc (MycERTM). This set up allowed two separate primary morphometric screens in MCF10A 3D cultures; one with and another without Myc oncogene challenge. These screens produced morphometric data from > 5,000 structures. The most interesting knockdown phenotypes were further analyzed with cell response markers (proliferation [i.e., Ki67], apoptosis [i.e., active caspase 3] and polarity change [i.e., α6-integrin, GM130]) and via 3D structures reconstructions obtained with confocal microscopy. The results from this screen for morphometric results have been published in Marques et al. (2016).
Materials and Reagents
- 24-well plates (VWR, catalog number: 391-3370 )
- Minisart filters 0.45 μm pore size (Sartorius, catalog number: 16537 )
- 6-well plates (VWR, catalog number: 700-1425 )
- 8-chamber slides (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 177402 )
- Microscope coverslips (MEDI PLAST FENNO, catalog number: 702-1051204 )
- Pipet tips
- 500 ml bottle
- Ultra adherent cell culture plates for 293ft cells
- Nunclon surface 10 cm (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 150350 )
- Nunclon surface 6 well plates (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 145380 )
- 293ft cells (Thermo Fisher Scientific, InvitrogenTM, catalog number: R70007 )
- MCF10A cells (ATCC, catalog number: CRL-10317TM )
- MCF10A-MycER cells (Nieminen et al., 2007)
- Plasmid and vectors:
- pCMV-dR8.91 (Delta 8.9) packaging plasmid (Marques et al., 2016)
- pCMV-VSVg envelope construct (Marques et al., 2016)
- pDSL_UGIH lentiviral shRNA vector (Alliance for Cellular Signaling) (Marques et al., 2016)
- pLKO lentiviral vector (Broad Institute TRC library, MISSION TRC-Hs 1.0 library) (Sigma-Aldrich) (Marques et al., 2016)
- pGIPZ mir-30 based vector (Open Biosystems) (Marques et al., 2016)
- Non-targeting shRNA control constructs (in pDSL_UGIH, pLKO.1 and pGIPZ lentiviral backbone) (Marques et al., 2016)
- Modified entry vector pENTR-H1 (Nieminen et al., 2007; Marques et al., 2016)
- Cell culture and transfection
- Dulbecco’s modified Eagle medium (DMEM) (Thermo Fisher Scientific, GibcoTM, catalog number: 21041-025 )
- Mammary epithelial basal medium (MCDB 170) (BioConcept AG Costumer made product based on USB powder formulation [Labome, catalog number: M2162 ] without L-glutamine; catalog number: 9-02S77-I)
- Transfection reagent - jetPEI (Polyplus, catalog number: 101-01N )
- Supplements for DMEM media (added before use; See Recipes):
- Fetal calf serum (FCS) (Biowest, catalog number: S1810-500 )
- L-glutamine (Lanza, catalog number: 17-605E )
- Penicillin-streptomycin (Lanza, catalog number: 17-602E )
- Phosphate buffered saline (PBS) (Sigma-Aldrich, catalog number: P4417 ) (Bionordika, catalog number: 17-516F/12 )
- Polybrene (Sigma-Aldrich, catalog number: H9268 )
- Matrigel® (Corning, catalog number: 356230 )
- RNeasy Mini Kit (Qiagen, catalog number: 74104 )
- ELB Lysis Buffer reagents (see Recipes)
- HEPES (Sigma-Aldrich, catalog number: H-3375 )
- EDTA (Sigma-Aldrich, catalog number: E5134 )
- Nonidet® P40 substitute (NP-40) (Sigma-Aldrich, catalog number: 74385 )
- Protease inhibitor cocktails
- Complete Mini (Roche Diagnostics, catalog number: 04693159001 )
- PhosphoStop (Roche Diagnostics, catalog number: 04906837001 )
- 0.05% trypsin (diluted from 5% trypsin-EDTA [10x]) (Thermo Fisher Scientific, GibcoTM, catalog number: 15400-054 )
- 4% paraformaldehyde phosphate buffer stock solution (PFA) (used as diluted 2%) (from powder: Sigma-Aldrich, catalog number: 16005-1Kg-R or from 20% aqueous solution Electron Microscopy Sciences, catalog number: 15713-S )
- Triton X-100, used as 0.25% dilution in PBS (Sigma-Aldrich, catalog number: 9002-93-1 )
- Blocking solution (see Recipes)
- Immunofluorescence (IF) buffer reagents (see Recipes)
- Sodium azide (NaN3) (Sigma-Aldrich, catalog number: S-2002 )
- Bovine serum albumin (BSA) (Biowest, catalog number: P6154 )
- Tween 20 (Sigma-Aldrich, catalog number: P9416 )
- Normal goat serum, used in 10% concentration (Thermo Fisher Scientific, GibcoTM, catalog number: PCN5000 )
- Counterstaining and Antibodies:
- Hoechst 33258 (Sigma-Aldrich, catalog number: 861405 )
Note: This product has been discontinued and it has been replaced by bisBenzimide H 33258 (Sigma-Aldrich, catalog number: B2883 ). - β-catenin (BD, catalog number: 610153 )
- GM-130 (BD, catalog number: 610823 )
- Anti-CD49f/α6-integrin (EMD Millipore, catalog number: CBL458 )
- E-cadherin (BD, catalog number: 610182 )
- Ki-67 (Leica Biosystems, catalog number: NCL-Ki67p )
- Active caspase-3 (Cell Signaling Technology, catalog number: 9661L )
- ZO-1 (Abcam, catalog number: ab59720 )
- Desmoplakin I+II (Abcam , catalog number: ab16434 or ab71690 )
- Desmoglein 2 (Abcam, catalog number: ab14415 )
- Power SYBR Green Cells-to-CT Kit (Thermo Fisher Scientific, AmbionTM, catalog number: 4402953 )
- Immu-MountTM reagent (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 9990412 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: 31434-5KG-R )
- Supplements for MCDB 170 media (added before use; See Recipes):
- Bovine pituitary extract (BPE) (Sigma-Aldrich, catalog number: P-1476 ) when not available use BPE (150 mg/batch) from Upstate (Sigma-Aldrich, catalog number: 02-104 )
- Epithelial growth factor (EGF) (Sigma-Aldrich, catalog number: E-9644 )
- Transferrin (Sigma-Aldrich, catalog number: T-2252 )
- Isoproterenol (Sigma-Aldrich, catalog number: I-5627 )
- Hydrocortisone (Sigma-Aldrich, catalog number: H-4001 )
- Insulin (Sigma-Aldrich, catalog number: I-9278 )
- Amphotericin B (Sigma-Aldrich, catalog number: A-2942 )
- Gentamicin (Sigma-Aldrich, catalog number: G-1397 )
- 100 nM 4-hydroxytamoxifen used to activate MycERTM construct (4-OHT; diluted from 1 mM stock) (Sigma-Aldrich, catalog number: H7904 )
- SuperScript Vilo cDNA Synthesis Kit (Thermo Fisher Scientific, catalog number: 11754-050 )
Equipment
- 37 °C, 5% CO2 incubator (Thermo Fisher Scientific, Thermo ScientificTM, model: FormaTM Series II 3110 )
- Shaker (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 4625Q )
- Confocal laser scanning Microscope (Zeiss, models: LSM Meta 510 and 780 equipped with argon [488], helium-neon [543 and 633] and diode [405] lasers and Plan-Neofluar 40x DIC objective [NA = 1.3, oil])
- Zeiss Axio vert 200 microscope equipped with Apotome system (Zeiss) and 20x Plan apochromat objective (NA = 0.8, air), MRr digital camera and Axiovision 4.4 software.
- Light Cycler 480 II instrument (Roche Diagnostics, model: Light Cycler 480 II )
- Nanodrop (Thermo Fisher Scientific, Thermo Scientific, model: NanoDrop 8000 )
- Heraeus Multifuge 3SR Plus (DJB Labcare, catalog number: 75004371 ) with Swing Swing-out rotor 4 place (DJB Labcare, catalog number: 75006445 ) and Microtitre carrier for 4 microtitre plates (DJB Labcare, catalog number: 75006449 )
Software
- ImageJ software (National Institute of Health, version 1.50i)
Procedure
- Constructing shRNA library to target putative human Epithelial Integrity Regulating genes (hEIR)
- shRNAs were designed to target the first and/or longest ENSEMBL transcript of each hEIR gene and 1-3 shRNA target sequences were chosen per transcript.
- Altogether, 136 shRNA oligos corresponding to the selected optimal target sequences (1-3 per gene) were cloned first into a pENTR cloning vector and thereafter to pDSL_UGIH lentiviral shRNA vector. Additionally 40 shRNA in pLKO lentiviral vector and 43 shRNAmirs in the pGIPZ mir-30 based vector were acquired to fully cover the previously selected target genes. Further information on the shRNA library designed to target epithelial integrity regulating genes can be found in Marques et al. (2016).
- Production of lentiviral particles
- Lentiviral vectors are transfected to 293ft cells growing in ultra adherent 24-well plates. 200,000 cells in 1 ml DMEM per well seeded one day prior transfections. Details below:
- Lentiviral vector mix: selected transfer plasmid (shRNA expression vector): Delta 8.9 (packaging): pCMV-VSVg (envelope) are mixed in a weight concentration ratio of 4:3:2 (tot. 10 μg; 4.44 μg, 3.33 μg, 2.22 μg) in 25 μl of 150 mM NaCl and incubated for 5 min at room temperature.
- Transfection: 1 μl JetPEI is mixed with 25 μl of 150 mM NaCl, and incubated for 5 min at room temperature. Lentiviral vector mix and transfection reagent JetPEI are mixed and incubated for 20 min at room temperature and dropwise added to the cells. The volume of media at the time of transfection is 0.5 ml.
- Transfected cells are incubated for at least 4 h at 37 °C/5% CO2 after which 0.5 ml of fresh media is added per well, making a total volume of 1 ml.
- After 72 h of incubation, viruses are harvested by filtering the media through a 0.45 μm filter.
- Occasionally viruses are produced in 6-well plates and the following formula is used: 293ft 1 million cells, 100 μl of 150 mM NaCl and 6 μl of JetPEI.
- MCF10A cells lentiviral infection
- MCF10A cells are seeded in 6-well plates aiming for a 50-60% confluence (3-4 x 105 cells per well) with 2 ml of MCDB170 + supplements media.
- Following day, media is changed and cells are washed with PBS. Afterwards medium and freshly harvested virus is added to cells in the ratio 1:1 (in a 6-well plate format add 500 μl viral suspension to 500 μl media).
- Polybrene is added to the cells at a ratio of 1:1,000 (8 μg/μl).
- After incubating the cells with the viruses and polybrene for 10 min at 37 °C, plates are centrifuged at 770 x g for 30 min at room temperature for virus transduction improvement.
- Cells are further incubated for a minimum of 4 h, after which media containing virus particles is to be replaced by 2 ml of fresh media.
- Cell collection for assays
72 h post-infection MCF10A cells are either lysed for RT-qPCR and/or Western blot analysis for knockdown efficiency analysis or seeded into Matrigel® for 3D culture assays. - shRNA knockdown validation
- RT-qPCR: Cells are lysed using the buffer supplied in the RNeasy kit and subsequent RNA isolation is performed using the kit buffers and instructions.
- Western blot: Cells are lysed in 40 μl of ELB lysis buffer supplemented with protease inhibitors (1/2 tablet [Complete Mini and PhosphoStop] per 10 ml of buffer).
- Setting up 3D culture for shRNA transduced MCF10A cells
- Matrigel® should be thawed overnight at 4 °C on ice.
- 8-chamber slides, pipets and pipet tips should be precooled at 4 °C for at least 1 h before starting the experiment.
- Each chamber is coated with 31 μl of Matrigel at 4 °C. Matrigel should be applied in a concentric way, avoiding contact with the chamber walls and the formation of bubbles.
- Matrigel-coated slides are incubated for 20-30 min at 37 °C to solidify the Matrigel.
- While Matrigel solidifies, shRNA transduced MCF10A growing cells should be trypsinized, counted and evaluated for viability. 1,500 viable cells are seeded per chamber on top of the Matrigel. Cells can be diluted in 20 μl to 400 μl of media, depending on the original concentration of viable cells in suspension.
- Media should be refreshed to the 3D cultures every third day, by carefully aspirating the old media and pipetting 400 μl of new media into the individual chambers.
- Fixing 3D cultured cells
- Media is removed and cells are washed twice with 400 μl of PBS.
- 200 μl of 2% PFA is used to fix the cells for 20 min at room temperature.
- After fixation, cells are washed twice with 400 μl of PBS at room temperature.
- If 3D cultures are not immunostained on the same day, cells can be stored for a couple of days (maximum time advised would be one week). This should be done at 4 °C, keeping the cells in the last PBS wash and using the 8-chamber slides lids to avoid dehydration of the samples.
- Immunostaining of the 3D cultures
- 3D cultures are permeabilized in their chambers for 10 min at room temperature using 200 μl of 0.25% Triton X-100 in PBS.
- Permeabilized 3D cultures are incubated at room temperature with 200 μl of blocking solution (immunofluorescence [IF] buffer with 10% normal goat serum) for 1-1.5 h.
- Primary antibodies are diluted in the blocking solution and added in 150 μl volume to each 3D cultures chambers (Antibodies concentration ranges from 1:50 to 1:500, see Notes for further information).
- Primary antibodies are incubated overnight at 4 °C using the 8-chamber slides lids.
- Cells are washed for three times with 400 μl of IF buffer at room temperature. Each wash performed for 20 min in a rocker at moderate speed.
- Afterwards cells in 3D cultures are incubated at room temperature for 45 min in 150 μl of the appropriate Alexa Fluor secondary antibody diluted in the blocking solution (In a concentration ranging from 1:150 to 1:1,000, see Notes for further information).
- 3D cultures are subsequently washed two times with 400 μl of IF buffer at room temperature.
- Nuclei are counterstained for 15 min with 150 μl of Hoechst 33258 (1:10,000 in PBS) and washed one more time with 400 μl of IF buffer at room temperature.
- After immunostainings, the 3D cultures are mounted by carefully removing the chamber and glue keeping them attached to the microscope slide. The coverslip is mounted with few drops of Immu-Mount reagent.
- Imaging of 3D cultures
- The formation of MCF10A structures should be monitored with a light microscope during the 3D cultures development (every other day is advised).
- Images for the morphometric analysis are acquired with an inverted epifluorescence microscope coupled with apotome system equipped with a 20x Plan objective (Figure 2).
- Cellular response (for example, Ki67, active caspase 3) and epithelial integrity markers (for example, α6-integrin, GM130, Desmoglein, Desmoplakin ZO-1) should be studied with higher resolution imaging using confocal microscopy. For further high-resolution morphological studies and 3D reconstructions, Z-stacks can be acquired from MCF10A structures and assembled into videos and 3D deconvolutions (Videos 1 and 2).Video 1. Animated visualization of an acinar structure Z-stack (top to bottom). In this example the MCF10A acinar structure was grown for 10 days after knockdown of DVL3 gene. Stained for E-cadherin in red and nuclei in blue.Video 2. 3D structure reconstruction from the previously shown Z-stack
- Analyses of the images
- Open images using ImageJ software for morphometric analyses.
- Use software Image menu to convert images to 16-bit type.
- Image threshold should be manually adjusted (Image menu, adjust submenu). Threshold protocol allows one to enhance the binary contrast to better visualize the borders of the structures for computational quantifications. Visual side-by-side inspection of the original and thresholded images is essential to validate the authentic character of processed images (Figure 2).
- If the thresholding process fuses neighboring 3D structures, a Pen tool should be used to separate shapes that are obviously individual shapes in the original image.
- The images are analyzed for the morphometric parameters area and circularity. These and other shape descriptors can be found under analyze menu, set measurements submenu.
- At least 30 structures per genotype should be analyzed for the shape parameters (area and circularity). Example data are shown in Figure 3.
- The protocols for confocal image analyses of different cellular responses and epithelial integrity alterations are described in Marques et al. (2016).
Representative data
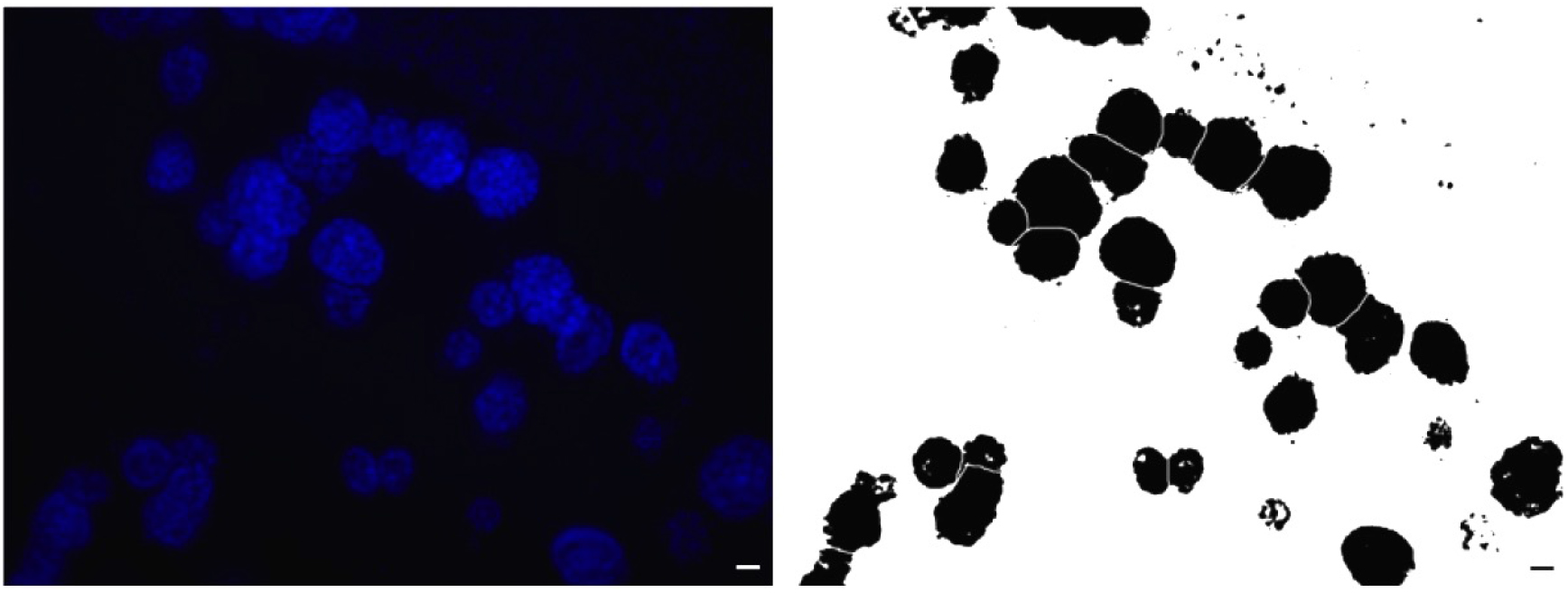
Figure 2. Example image before (left) and after (right) thresholding the nuclear stained image for morphometric analysis (Scale bars = 20 μm)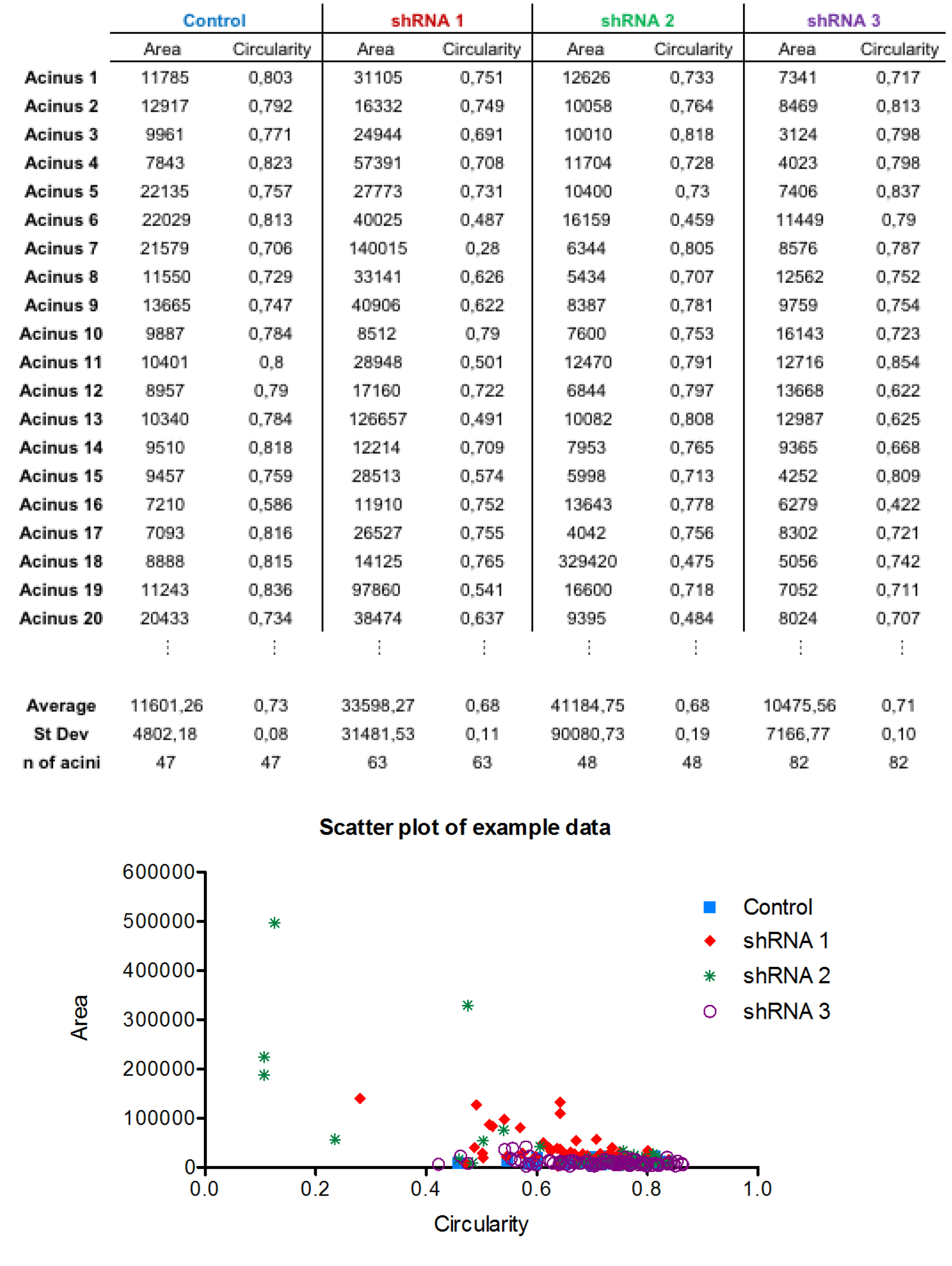
Figure 3. Example of the morphometric quantifications and data visualization with scatter plot. In this set of experiments, shRNA3 (purple; PARD6B shRNA) induced mild alterations in the area and circularity of the 3D acinar structures. In contrast, both shRNA1 (red; PARD6G shRNA) and shRNA2 (green; DVL3 shRNA) decreased circularity (symmetry) and increased the area (size) of the 3D acinar structures. Therefore, DVL3 and PARD6G, but not PARD6B appear to be important for the development of proper epithelial integrity.
Data analysis
- The morphometric values corresponding to each construct are averages of a minimum of 30 3D MCF10A structures analyzed from > 3 fields-of-view (FOV) per each experiment.
- The morphometric average for the control shRNA transduced structures is set to 1 and the phenotypic data can be analyzed as fold changes relative to the control.
- The selected thresholds were set to 20% fold change in epithelial structure area and > 10% in symmetry. For more details about the data analysis, see Marques et al. (2016).
- Based on the threshold values, the shRNA caused phenotypes were grouped according to the changes in size and symmetry (for example, large/hyperplastic, small & asymmetric, etc.). The results of the screen and shRNA induced phenotypes are described in Marques et al. (2016).
Notes
- An important methodological concern is the variability between Matrigel lots. Different lots can have dramatically different bioactivity, affecting the viability, morphology and timing of maturation (cell cycle arrest). Laboratories working with Matrigel based 3D cultures should have an internal quality control system to rigorously test each new lot for 3D structure growth supporting bioactivity for the different cell lines to be used.
- Primary and secondary antibody concentrations need to be tested in preliminary experiments. The ratios provided in the protocol are only guidelines.
Recipes
- MCDB170 supplements added per 500 ml bottle
2.5 ml BPE from stock (200x stock - 14 mg/ml; final concentration of 70 μg/ml)
25 μl EGF from stock (final concentration of 5 ng/ml)
250 μl transferrin from stock (10 mg/ml 2,000x stock; final concentration of 5 μg/ml)
1 ml isoproterenol from stock (500x stock; final concentration of 10 µM)
250 μl hydrocortisone from stock (2,000x stock; final concentration of 0.5 μg/ml)
250 μl insulin from stock (2,000x stock; final concentration of 5 μg/ml)
5 ml amphotericin B from stock (50 μg/ml)
500 μl gentamicin from stock (50 μg/ml) - DMEM supplements added to 500 ml bottle
10% FCS (50 ml from stock)
2.5 ml L-glutamine from 2 mM stock
1% penicillin-streptomycin (5 ml from stock) - ELB lysis buffer
150 mM NaCl
50 mM HEPES, pH 7.4
5 mM EDTA
1% NP-40 - Immunofluorescence (IF) buffer
7.7 mM NaN3
0.1% bovine serum albumin
0.2% Triton X-100
0.05% Tween 20
in PBS - Blocking solution
10% goat serum in IF buffer
Acknowledgments
We would like to acknowledge all the Klefström laboratory members for the critical advice and input in developing this protocol. Special thanks to lab members Johanna Englund, Topi Tervonen, Anne Makelä and Minna Ahvenainen for setting up many methods described here, Tiina Raatikainen and Tarja Välimäki for technical assistance and Eric Coles for critical comments and language accuracy. Biomedicum Imaging Unit and Biomedicum Functional Genomics Unit are acknowledged for core services and technical support. The original protocols for MCF10A 3D culture were described in Debnath et al. (2002). These protocols were modified to further expand genetic approaches (Partanen et al., 2007; Marques et al., 2016). The shRNA screen to identify epithelial regulating genes in MCF10A 3D culture has been published in Marques et al. (2016). This study was funded by the Academy of Finland, TEKES, Finnish Cancer Organizations, Helsinki Graduate Program in Biotechnology and Molecular Biology and Innovative Medicines Initiative Joint Undertaking under grant agreement No. 115188.
References
- Debnath, J. and Brugge, J. S. (2005). Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer 5(9): 675-688.
- Debnath, J., Mills, K. R., Collins, N. L., Reginato, M. J., Muthuswamy, S. K. and Brugge, J. S. (2002). The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell 111(1): 29-40.
- Marques, E., Englund, J. I., Tervonen, T. A., Virkunen, E., Laakso, M., Myllynen, M., Makela, A., Ahvenainen, M., Lepikhova, T., Monni, O., Hautaniemi, S. and Klefstrom, J. (2016). Par6G suppresses cell proliferation and is targeted by loss-of-function mutations in multiple cancers. Oncogene 35(11): 1386-1398.
- Nieminen, A. I., J. I. Partanen, A. Hau and J. Klefstrom (2007). c-Myc primed mitochondria determine cellular sensitivity to TRAIL-induced apoptosis. EMBO J 26(4): 1055-1067.
- Partanen, J. I., Nieminen, A. I., Makela, T. P. and Klefstrom, J. (2007). Suppression of oncogenic properties of c-Myc by LKB1-controlled epithelial organization. Proc Natl Acad Sci U S A 104(37): 14694-14699.
- Partanen, J. I., Tervonen, T. A. and Klefstrom, J. (2013). Breaking the epithelial polarity barrier in cancer: the strange case of LKB1/PAR-4. Philos Trans R Soc Lond B Biol Sci 368(1629): 20130111.
- Rejon, C., Al-Masri, M. and McCaffrey, L. (2016). Cell polarity proteins in breast cancer progression. J Cell Biochem.
- Roignot, J., Peng, X. and Mostov, K. (2013). Polarity in mammalian epithelial morphogenesis. Cold Spring Harb Perspect Biol 5(2).
- Shamir, E. R. and Ewald, A. J. (2014). Three-dimensional organotypic culture: experimental models of mammalian biology and disease. Nat Rev Mol Cell Biol 15(10): 647-664.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Marques, E. and Klefström, J. (2016). Lentiviral shRNA Screen to Identify Epithelial Integrity Regulating Genes in MCF10A 3D Culture. Bio-protocol 6(23): e2050. DOI: 10.21769/BioProtoc.2050.
Category
Cancer Biology > General technique > Cell biology assays > Cellular transformation
Cell Biology > Cell-based analysis > Gene expression
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










