- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Vascular Smooth Muscle Cell Isolation and Culture from Mouse Aorta
Published: Vol 6, Iss 23, Dec 5, 2016 DOI: 10.21769/BioProtoc.2045 Views: 23424
Reviewed by: Jia LiGuillermo GomezAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Selective Enrichment and Identification of Cerebrospinal Fluid-Contacting Neurons In Vitro via PKD2L1 Promoter-Driven Lentiviral System
Wei Tan [...] Qing Li
Nov 20, 2025 1343 Views

Revisiting Primary Microglia Isolation Protocol: An Improved Method for Microglia Extraction
Jianwei Li [...] Guohui Lu
Dec 5, 2025 1475 Views

Optimization of Adipogenic Differentiation Protocol for Murine and Human Cell Culture Models
Junwan Fan [...] Wenyan He
Jan 20, 2026 230 Views
Abstract
Vascular smooth muscle cells (SMC) in the ascending thoracic aorta arise from neural crest cells, whereas SMCs in the descending aorta are derived from the presomitic mesoderm. SMCs play important roles in cardiovascular development and aortic aneurysm formation. This protocol describes the detailed process for explanting ascending and descending SMCs from mouse aortic tissue. Conditions for maintenance and subculture of isolated SMCs and characterization of the vascular SMC phenotype are also described.
Keywords: Tissue cultureBackground
Vascular smooth muscle cells (SMCs) make up the muscular medial layer of arteries. Larger elastic arteries, such as the aorta, have multiple concentric lamellae consisting of aligned smooth muscle cells sandwiched between elastin fibers. The elastin and collagen present within the medial layer of elastic arteries allow it to distribute the force generated by the heart throughout the vessel wall (Wagenseil and Mecham, 2009). Smaller muscular arteries, by contrast, have only an internal and external elastic lamina bounding the smooth muscle layer. These arteries are downstream in the arterial tree and thus bear less force from blood flow.
Vascular smooth muscle cells, unlike cardiac and skeletal muscle cells, are capable of modulating their phenotype in response to vascular injury or environmental cues. Under normal physiologic conditions, quiescent, contractile SMCs populate the artery wall and contract to regulate vascular tone and keep blood flow continuous in response to pulsatile pressures. Contractile SMCs are characterized by high expression of smooth muscle-specific contractile genes, including smooth muscle specific α-actin and myosin heavy chain (Owens et al., 2004). However, in response to injury or mitogenic stimuli, SMCs downregulate expression of the contractile genes and take on a synthetic phenotype: they proliferate rapidly, migrate into the site of injury, and remodel the extracellular matrix by synthesizing both matrix-digesting enzymes and new matrix proteins. Many vascular diseases are associated with a synthetic SMC phenotype, including atherosclerosis (Owens, 1995).
SMCs located in different areas of the body actually arise from diverse embryonic lineages (Majesky, 2007). For example, the SMCs populating the ascending thoracic aorta and the cerebrovasculature are derived from neural crest cells. However, SMCs in the descending thoracic aorta come from mesodermal origins. These distinctions affect the ultimate properties of the SMCs, so it is important when designing experiments to use SMCs from the same developmental origin where the phenotype of interest arises.
In this protocol, we give detailed instructions for isolating and culturing SMCs from the ascending and descending thoracic aortas in the mouse. We have previously used this technique to isolate SMCs from genetically modified mice and their wild-type littermates to provide an in vitro system for looking at the effect of genetic changes on SMC phenotype (Cao et al., 2010; Kuang et al., 2012; Kuang et al., 2016; Papke et al., 2013; Kwartler et al., 2014).
Materials and Reagents
- For initial explant
- Dissection material: 2 sterile forceps, 2 sterile tweezers, 1 sterile scissor, 2 sterile scalpels
- Disposable scalpels (Aspen Surgical, catalog number: 371621 )
- Four 60 mm tissue culture dishes per sample (Corning, catalog number: 430589 )
- Syringe, 10 ml
- Syringe filter, 0.22 µm pore
- 500 ml filtration unit, 0.22 µm pore (EMD Millipore, catalog number: SCGPU01RE )
- Parafilm
- Aluminum foil
- 2.5% avertin-2,2,2-tribromoethanol (Sigma-Aldrich, catalog number: T48402-25g ) arrives as powder, make 100% stock solution by dissolved 25 g 2,2,2-tribromoethanol in 25 ml of 2-methyl-2-butanol (Sigma-Aldrich, catalog number: 240486-100ML ), then dilute to 2.5% solution in sterile water. Both the stock solution and the dilution are light sensitive, so should be stored in the dark at 4 °C
- 70% ethanol
- Dulbecco’s phosphate buffered saline (DPBS) with calcium and magnesium (GE Healthcare, HycloneTM, catalog number: SH30028.02 )
- Components of aorta biopsy storage media:
- Waymouth’s MB 752/1 medium, 500 ml (Thermo Fisher Scientific, GibcoTM, catalog number: 11220035 )
- Antibiotic-antimycotic, 100x (Sigma-Aldrich, catalog number: A5955 )
- L-glutamine, 100x (Sigma-Aldrich, catalog number: G7513 )
- Sodium bicarbonate (Sigma-Aldrich, catalog number: S8761 )
- MEM non-essential amino acids (Sigma-Aldrich, catalog number: M7145 )
- HEPES buffer (Sigma-Aldrich, catalog number: H0887 )
- Collagenase type 1 (Sigma-Aldrich, catalog number: C1639 )
- Elastase, Pancreatic type 1 from porcine pancreas (Sigma-Aldrich, catalog number: E1250 )
- Soybean trypsin inhibitor (Thermo Fisher Scientific, GibcoTM, catalog number: 17075-029 )
- Heat inactivated fetal bovine serum (FBS) (Atlanta Biologicals)
- SmBm bullet kit (Lonza, catalog number: CC3182 )
- FGF
- EGF
- Insulin
- Gentamicin and included FBS - Do not use!
- For continued culture
- 500 ml filtration unit, 0.22 µm pore (EMD Millipore, catalog number: SCGPU01RE )
- Freeze vials (2 ml)
- Fetal bovine serum (FBS, Atlanta Biologicals)
- Antibiotic-antimycotic, 100x (Sigma-Aldrich, catalog number: A5955 )
- SmBm bullet kit (Lonza, catalog number: CC3182 )
- FGF
- EGF
- Insulin
- FGF
- TrypLE express (Thermo Fisher Scientific, GibcoTM, catalog number: 12604013 )
- DMSO
- For immunofluorescence to confirm SMC identity
- Coverslips (UV treated, please see Data analysis section below for details)
- 6 well plates
- Hemocytometer
- 3-6 mice, preferably aged 4-6 weeks old
- SmBm basal media (Lonza, catalog number: CC3181 )
- Fetal bovine serum (FBS) (Atlanta Biologicals)
- Recombinant human TGF-β1 (rhTGF- β1, R&D Systems)
- 16% formaldehyde (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 28906 ). Dilute 1 to 4 to a final concentration of 4% in DPBS with calcium and magnesium
- Blocking buffer (0.3% Triton X, 0.5% BSA in DPBS)
- DPBS
- Smooth muscle α-actin antibody (Sigma-Aldrich, catalog number: A5228 )
- Anti-mouse secondary antibody conjugated with fluorescence
- Mounting medium with DAPI (Vectashield, catalog number: H-1200 )
- Aorta biopsy storage media (see Recipes)
- Complete smooth muscle media (see Recipes)
- Smooth muscle cell freeze media (see Recipes)
- Digestive enzyme mix (see Recipes)
Equipment
Note: The reproducibility of the experiment is not dependent on the specific brand or model of these equipment. Any microscope, tissue culture hood/incubator, centrifuge, etc., will give similar results.
- Magnetic stirrer
- Dissecting microscope
- Sterile tissue culture hood
- Sterile tissue culture incubator
- 10 ml pipet
- Water bath
- Tabletop centrifuge
- 200 µl pipette
- T25 flasks
- T75 flasks
- Slow freeze container (either a foam container or an isopropanol based container will work)
- Hemocytometer
- Inverted microscope
- Hot bead sterilizer
Procedure
- Preparation of reagents
- To make aorta biopsy storage media, add the following to the bottle of 500 ml. Waymouth’s MB 752/1 medium.
- 5 ml of antibiotic/antimycotic
- 6.25 ml of L-glutamine
- 15 ml of sodium bicarbonate
- 5 ml of MEM non-essential amino acids
- 5 ml of HEPES buffer
- 5 ml of antibiotic/antimycotic
- Preparation of avertin solution
- Add 15.5 ml tert-amyl alcohol to 25 g avertin (2-2-2-tribromoethanol) and stir the solution on a magnetic stirrer until avertin is completely dissolved. This will probably take overnight. The stock should be stored in a dark bottle at room temperature and capped tightly.
- Combine 0.5 ml avertin stock and 39.5 ml normal saline or PBS in a 50 ml graduated cylinder.
- Drop in magnetic stir bar, seal graduate cylinder with Parafilm and completely wrap cylinder with aluminum foil to exclude all light.
- Stir overnight to dissolve.
- Filter the working solution through 0.2 micron filter into a dark bottle. Keep working solution at 4 °C.
- Stock and working solution containers will be properly labeled (name, date made, concentration and initials of lab member).
- Stock solution should be discarded after four months, and working solution should be discarded after 2 weeks.
- All solutions should not be used if a precipitate or discoloration is detected.
- To make complete smooth muscle media (SmBm complete), filter the following through a 0.22 µm vacuum filter and add to a bottle of 500 ml SmBm basal media from Lonza. This media can be stored at 4 °C for up to 2 months or until the expiration date from the manufacturer on the bottle, whichever is sooner. It is important to note that the aliquot of gentamicin contained in the SmBm bullet kit should not be added.
- 100 ml FBS
- Aliquot of FGF contained in the bullet kit (1 ml).
- Aliquot of EGF contained in the bullet kit (0.5 ml).
- Aliquot of insulin contained in the bullet kit (0.5 ml).
- 5 ml of antibiotic/antimycotic
- 100 ml FBS
- Preparation of digestive enzymes and other reagents
- Thaw and filter one 500 ml bottle of FBS and store at 4 °C in 50 ml aliquots.
- To make smooth muscle cell freeze media, combine 45 ml of SmBm complete with 5 ml filtered FBS and 5 ml sterile DMSO.
- Collagenase type 1 is received as a powder. The final concentration needed for a 3-4 h digestion is 1.00 mg/ml. To prepare the stock solution, add 2.5 ml aorta biopsy storage media to re-suspend the 50 mg collagenase. Mix well. Make 500 µl aliquots and store at -20 °C.
- Soybean trypsin inhibitor is received as a powder. The final concentration needed for a 3-4 h digestion is 0.25 mg/ml. To prepare the stock solution, take 2.5 mg and dissolve in 10 ml aorta biopsy storage media, then make 10 aliquots, 1 ml/aliquots
- Elastase type 1 is received as a liquid. The bottle can be stored at 4 °C. Do not aliquot as the reagent is not stable when aliquoted and stored at -20 °C. The final concentration needed for a 3-4 h digestion is 0.1875 mg/ml.
- To make aorta biopsy storage media, add the following to the bottle of 500 ml. Waymouth’s MB 752/1 medium.
- Dissection of the mouse to remove aortic tissue
- Anesthetize the experimental mice (3-6 mice/explant) by intraperitoneal (IP) injection with 2.5% avertin. Use 0.5 ml of solution for each mouse. To determine when anesthesia is effective, pinch the toe of the mouse hard between your nails. If the mouse kicks or reacts at all, the anesthesia is incomplete. Wait a couple of additional minutes and test again until there is no response to the toe pinch. If anesthesia is still incomplete after 5-7 min you can dose the mouse again with an additional 0.3 ml of avertin.
- Place the mouse under a light source for dissection.
- Cut the skin of the mouse from the abdomen to the top of the thorax.
- Lift the sternum with tweezers and cut the diaphragm. Then cut away the lower part of the ribcage to expose the heart, remove the lungs and thymus, and cut away the esophagus, expose the aorta running along the spine. See Figure 1 and Video 1.
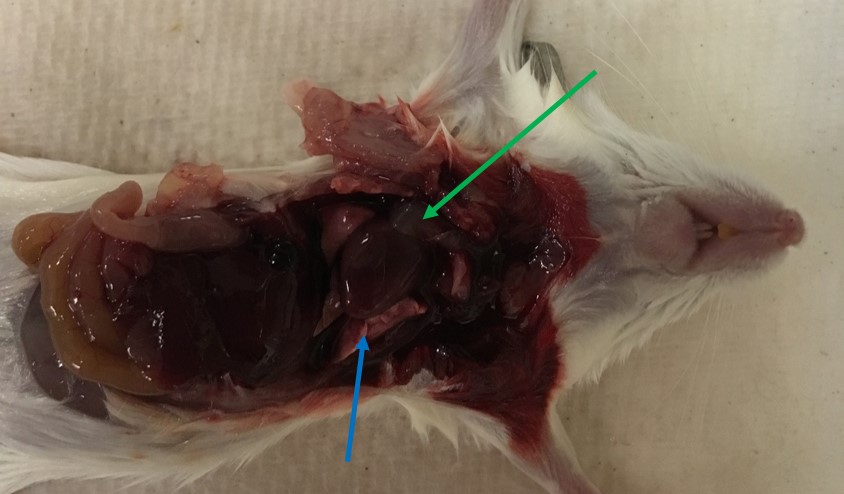
Figure 1. Mouse thoracic cavity. Picture shows the mouse after chest has been opened. Blue indicates the lung tissue and green the thymus tissue which must both be removed to give better access to the heart and aorta.Video 1. Opening of mouse thoracic cavity and removal of tissues. This video shows the first part of the dissection from the opening of the mouse thoracic cavity to the removal of the lungs and trachea. - Place the mouse under a dissecting microscope.
- Remove the remaining tissues and use microdissection scissors or forceps to remove all of the fat around the top of the heart, the ascending aorta, and its branches. See Figure 2.
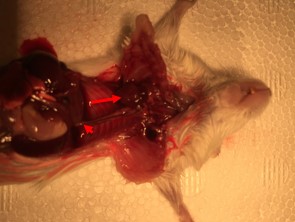
Figure 2. Exposed heart and descending aorta in a mouse. Picture shows the mouse after chest has been opened and lungs, thymus, and other tissues have been removed. The red arrow indicates the heart, which should be pulled up slightly to expose the ascending aorta and carotid branches. The red arrowhead indicated the descending aorta which should be clipped below the diaphragm. - Gently pull up on the heart with forceps, without breaking the ascending aorta. Use forceps or scissors to gently separate the descending aorta from the spine. Clip the descending aorta just below the diaphragm. Clip the carotid arteries to release the ascending aorta. See Video 2. Put the heart, ascending, and descending aortas which are all connected in a 60 mm dish with sterile DPBS.Video 2. Removal of mouse heart and aorta. This video shows the second part of the dissection: removing the heart and aorta.
- Place the dish under the dissecting microscope and remove any fatty or connective tissues from the ascending and descending aorta. See Figure 3. Separate the ascending and descending aorta by making a cut just distal to the origin of the left common carotid artery. Put ascending and descending aorta separately in new dishes with DPBS labeled according to the region of the aorta and the genotype of the mouse.
- Working quickly but carefully, repeat for each experimental mouse until all the tissue has been separated. Pool ascending aortas and descending aortas for each explant.
- Move all the dishes with aortic tissue to the tissue culture hood.

Figure 3. Appearance of the dissected aorta. Whole mount image showing the aortic tissue after it has been removed from the mouse and completely cleaned. Ascending aorta is marked by a red arrow.
- Anesthetize the experimental mice (3-6 mice/explant) by intraperitoneal (IP) injection with 2.5% avertin. Use 0.5 ml of solution for each mouse. To determine when anesthesia is effective, pinch the toe of the mouse hard between your nails. If the mouse kicks or reacts at all, the anesthesia is incomplete. Wait a couple of additional minutes and test again until there is no response to the toe pinch. If anesthesia is still incomplete after 5-7 min you can dose the mouse again with an additional 0.3 ml of avertin.
- Explanting of the tissue
Note: Please note that all steps for the rest of the protocol should be carried out under a tissue culture hood using standard sterile tissue culture techniques.- Prepare the enzyme mix
- Thaw one aliquot each of collagenase and trypsin inhibitor at 4 °C before beginning the dissection of the mice.
- For each explant, use 5 ml of aorta biopsy storage media. If you are doing a 3-4 h digestion during the day, add 250 µl collagenase, 100 µl trypsin inhibitor, and 324 µl* elastase for each 5 ml of aorta biopsy storage media. For an overnight (15-18 h) digestion, use 1:10 dilution so 25 µl collagenase, 10 µl trypsin inhibitor, and 32.4 µl* elastase.
*Note: The amount of elastase to add was calculated based on the mass and volume of the bottle. If you order elastase from a different source, you will need to recalculate the amount of this enzyme to add.
- Thaw one aliquot each of collagenase and trypsin inhibitor at 4 °C before beginning the dissection of the mice.
- Pour 70% ethanol, DPBS, and aorta biopsy storage media into three separate 60 mm tissue culture dishes.
- Using forceps, take each isolated aorta from its storage tube and wash the piece of tissue in ethanol, then DPBS, then biopsy media. Do not leave the tissue in ethanol for more than about 10-15 sec, though leaving it in DPBS or media for longer is OK.
- Place each piece of tissue into the lid of a 60 mm tissue culture dish. Using a scalpel and forceps, cut tissue into small pieces-make sure to cut the tissue both circumferentially and longitudinally to increase the surface area exposed to the digestion mix. The pieces will be variable in size, but the average size should be 1-2 mm in diameter.
- Place the chopped up tissue pieces into a new 60 mm tissue culture dish containing the 5 ml of media plus digestive enzyme mix made up in step C1. Label the dish.
- Place dish into incubator (37 °C). If doing a 3-4 h digestion, check at 2 h and every 30 min thereafter. If doing an overnight digestion, leave overnight, check at 15 h and every hour thereafter.
- When the tissue looks mostly digested with several floating single cells, but still some large clumps of tissue, pipet up and down tissue and media 5-10 times using a 10 ml pipet to help release cells from tissue. Place back into incubator for 30 more minutes. Also, remove filtered FBS from fridge to warm up to room temperature and place complete SmBM media into water bath to warm up to 37 °C.
- At the end of incubation time, look at tissue under microscope to make sure that the cells are free from the tissue. See Figure 4. If the tissue has been well digested (there should still be some intact pieces of tissue), add 2.5 ml of Serum and pipet up and down ~5 times. Next add 2.5 ml of complete SmBM media and pipet up and down ~5 times.
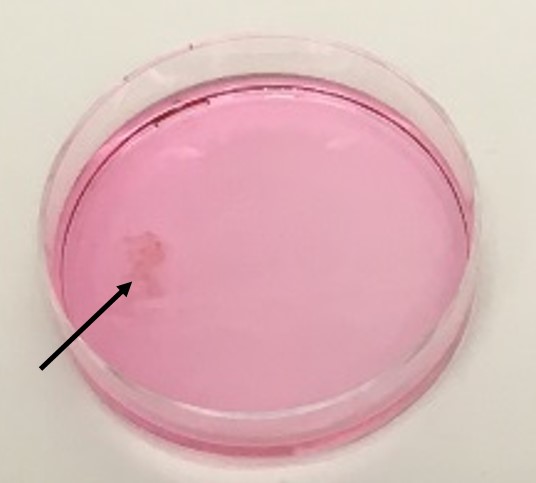
Figure 4. Appearance of the digested tissue. Photo of the dish after digestion, with some ‘net-like’ tissue floating in it (black arrow) but a majority of tissue pieces digested. - Transfer the entire contents of the tissue culture dish (should be 10 ml total) into one 15 ml tube.
- Spin down at 180 x g (about 700 rpm in a Sorvall Legend T+ tabletop centrifuge) at room temperature for 5 min.
- Carefully check to see if there is any floating tissue piece in the tube. If so, use a 200 µl pipette to remove the tissue piece and place it directly into a labeled T25 flask with complete SmBm. Once there are no tissue pieces floating, remove supernatant and resuspend the pellet in 5 ml of complete SmBM media.
Note: If the supernatant was cloudy, repeat steps C10 and C11 until the supernatant is clear. If the supernatant was clear, proceed to step C12. - Transfer the 5 ml complete SmBm with resuspended cells to a labeled T25 flask. Place flask in incubator.
- Allow cells to settle and adhere to the flask. Don’t change media for at least 48 h. If cells still haven’t settled down at this point, don’t remove any media and add 2 ml of media to the flask. Let the cells settle for another 1 to 2 days.
- Prepare the enzyme mix
- Maintenance and subculture of mouse vascular smooth muscle cells.
- Once SMCs grow up, feed the cells with fresh media every 3-4 days.
- Prewarm the SmBm complete media bottle in a 37 °C water bath for at least 15 min prior to feeding cells.
- Aspirate media from each flask and discard.
- Add 5 ml of media for each T25 flask without disturbing the cell layer.
- Return flasks to the incubator.
- When the cells reach about 70% confluent, or when some areas start to get very tightly packed and almost overgrown, the cells are ready to split. See Figure 5. Passage the SMCs from one T25 flask to one T75 flask.
- Prewarm the SmBm complete media bottle in a 37 °C water bath and allow the TrypLE Express reagent to equilibrate at room temperature for at least 15 min prior to splitting cells.
- Aspirate media from each flask and discard.
- Wash each flask once with 4 ml of DPBS, allow it to cover the cell layer, then aspirate and discard the DPBS.
- Add 1 ml TrypLE Express reagent to each flask. Make sure the trypsin is evenly covering the bottom of the flask/cell layer.
- Place the flask in the 37 °C incubator for 3 min.
- Confirm digestion under microscope. If most cells are round, bang the flask twice on both sides. If most cells are still attached, incubate at room temperature for 1 min and then confirm digestion and bang the flasks twice on both sides.
- Add 4 ml of SmBm complete to each flask to neutralize the trypsin enzyme. Pipet up and down to detach all the cells and completely neutralize the enzyme.
- Transfer all 5 ml of liquid plus cells to a 15 ml conical tube.
- Spin down 5 min at 180 x g in a tabletop centrifuge at room temperature.
- Aspirate supernatant, resuspend cell pellet in 10 ml of SmBm complete, and transfer all 10 ml to a labeled T75 flask. Mark the date of split and the passage number (P1 for this first passaging).

Figure 5. Appearance of confluent SMCs. The left picture shows a 100% confluent field of smooth muscle cells. These cells need to be passaged within 24 h. The right panel shows smooth muscle cells around 70% confluent. Note the spindle-shaped morphology which is characteristic of SMC cultures. - When the SMCs have expanded, the cell aliquots can be frozen. Follow steps D2a-D2i in the above protocol for passaging SMCs. One T75 flask can be split into 2 frozen cell aliquots. A confluent T75 flask should have around 2 million cells, so each freeze vial will have around 1 million cells. After spinning down, follow the below steps to freeze cells.
- Aspirate supernatant, resuspend cell pellet in 3 ml SMC freeze media (see Recipes).
- Transfer 1.5 ml of the freeze media with dissolved pellet to each of two cryovials. Cryovials should be labeled with the passage number, date of freezing, and cell line.
- Place cryovials into a slow freeze container (either a foam container or an isopropanol based container will work) and freeze in the -80 °C freezer overnight.
- Transfer the frozen aliquots to a liquid nitrogen storage system for long-term storage.
- Aspirate supernatant, resuspend cell pellet in 3 ml SMC freeze media (see Recipes).
- When you want to thaw your SMC aliquots, a vial of frozen cells can typically be thawed to one T75 flask.
- Prewarm SmBm complete media in the 37 °C water bath for at least 15 min prior to removing cells.
- Retrieve the cell vial from the liquid nitrogen freezer. Warm quickly by placing the cryovials in the 37 °C water bath for about 1-2 min.
- Add 8 ml of SmBm complete to a 15 ml conical tube. Then transfer the 1.5 ml of thawed cells into the same conical tube.
- Spin down for 5 min at 180 x g in a tabletop centrifuge at room temperature.
- Remove and discard supernatant, resuspend the cell pellet in 10 ml of SmBm complete and transfer to a T75 flask.
Data analysis
- Characterization of SMC phenotype
In order to confirm the identity of the SMCs, perform immunofluorescence for smooth muscle α-actin, which should be visible in filaments in a differentiated SMC. When the cells are 80% confluent, they are ready to plate. An hour before beginning to work with the cells, place coverslips into a 6-well plate (one coverslip per well, as many as you want to plate, for this experiment we recommend at least two) under the cell culture hood. Turn on the UV in the hood for at least 30 min to sterilize the coverslips. Then to begin, follow steps D2a-D2i for passaging cells above. After spinning down, aspirate supernatant and resuspend the cell pellet in a smaller volume-typically 2 ml of SmBm complete.- Use 10 µl of the resuspended cell mixture to count cells in a hemocytometer.
- Calculate the volume needed for 5,000 cells. Add the calculated volume of cell mixture to the well containing the coverslip and add additional SmBm complete media to each well up to 2 ml volume. Any remaining cells can be used for other experiments or put back into flasks as desired.
- 18-24 h after plating the cells, change media to low serum SmBm media, which drives differentiation of vascular SMCs. This is SmBM basal media with only 1% FBS and 1x antibiotic/antimycotic added (for 500 ml bottle of SmBm basal media, add 5 ml FBS and 5 ml of 100x antibiotic/antimycotic). Leave low serum SmBm on your cells at least 24 h.
- If desired, you can treat some coverslips with rhTGF-β1 to further induce differentiation. 24 h after switching to low serum SmBm media, add 10 ng/ml rhTGF-β1 to low serum SmBm media and add this media to the coverslips. Leave rhTGF-β1 containing media on the cells for 48 h.
- When the treatment is done, remove media, wash the cells once with DPBS, and add 1 ml of 4% paraformaldehyde solution to each well to fix the cells. Store at 4 °C until the cells are ready to stain.
- For staining, use 50 µl droplets of liquid for each coverslip on Parafilm and invert the coverslip onto the droplet to save antibody. First, incubate the coverslips in blocking buffer for 1 h (blocking buffer is 0.3% Triton-X, 0.5% BSA in DPBS).
- Wash coverslips 3 times 5 min in DPBS
- Incubate coverslips in 1:500 anti-smooth muscle α-actin antibody made up in blocking buffer at 4 °C overnight.
- Wash coverslips 3 times 5 min in DPBS.
- Incubate coverslips in 1:500 fluorescent-conjugated anti-mouse IgG secondary antibody made up in blocking buffer for 3 h at room temperature.
- Wash coverslips 3 times 5 min in DPBS
- Mount coverslips onto microscope slides with mounting media with DAPI (nuclear counterstaining, Vectashield).
- Use nail polish or other desired sealant to seal coverslip to the slide.
- Assess staining using desired fluorescent microscope. At least 95% of the cells should have smooth muscle α-actin filaments visible in the cells (Figure 6).
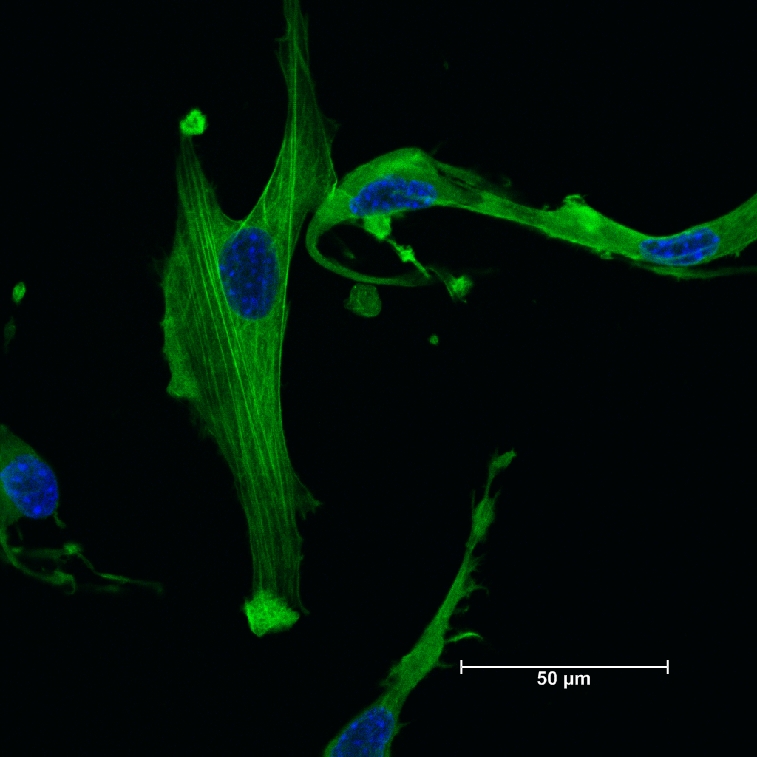
Figure 6. Actin filaments in isolated smooth muscle cells. Immunofluorescence staining for SM α-actin (green) shows polymerized filaments of SM α-actin. Nuclei are counterstained with DAPI (blue).
- Use 10 µl of the resuspended cell mixture to count cells in a hemocytometer.
Notes
- It is critical to correctly time the ethanol wash step (step C3 under ‘Explant’ protocol above). Too long a time in the ethanol will kill the cells and dramatically lower success. Too short a time in the ethanol wash may lead to bacterial contamination of the sample.
- Although the product numbers for the digestive enzymes listed above have been extensively tested in our hands, we have also had success with other versions of Collagenase and Elastase. The most critical factor is the concentration of active enzyme and the time of digestion. Any alternate enzyme will need to be tested for optimal time of digestion.
- Typically, in our hands primary smooth muscle cells retain their phenotype in culture for at least six passages. We always use low passage cells (< P6) for our experiments. However, the cells may be usable beyond passage 6 but we recommend assessing the smooth muscle cell phenotype of higher passage cells before use. Phenotype can be assessed by using the staining protocol described above and/or by expression of smooth muscle myosin heavy chain, encoded by Myh11, which is the most specific marker for differentiated smooth muscle cells.
Recipes
- Aorta biopsy storage media
500 ml of Waymouth’s MB 752/1 medium
5 ml of 100x antibiotic-antimycotic
6.25 ml of L-glutamine
15 ml of sodium bicarbonate
5 ml of MEM non-essential amino acids
5 ml of HEPES buffer - Complete smooth muscle media
500 ml of SmBm basal media
100 ml of FBS
1 ml of FGF contained in the bullet kit
0.5 ml of EGF contained in the bullet kit
0.5 ml of insulin contained in the bullet kit
5 ml of antibiotic/antimycotic - Smooth muscle cell freeze media
45 ml of complete smooth muscle media
5 ml of filtered FBS
5 ml of sterile DMSO - Digestive enzyme mix (for 3-4 h digestion time)
5 ml aorta biopsy storage media
250 μl collagenase
100 μl trypsin inhibitor
324 μl elastase
Acknowledgments
This work was supported by National Institutes of Health: NIH (RO1 HL62594 and P01HL110869-01), John Ritter Foundation, Vivian L. Smith Foundation, and Ehlers Danlos Syndrome Network CARES.
The method was published in Kuang et al. (2016) and it is an adaptation of the methods used in Gordon et al. (1986) and Lemire et al. (1994). Cells explanted using this method were also utilized in the following publications: Cao et al. (2010), Kuang et al. (2012), Kwartler et al. (2014), Papke et al. (2013).
References
- Cao, J., Gong, L., Guo, D. C., Mietzsch, U., Kuang, S. Q., Kwartler, C. S., Safi, H., Estrera, A., Gambello, M. J. and Milewicz, D. M. (2010). Thoracic aortic disease in tuberous sclerosis complex: molecular pathogenesis and potential therapies in Tsc2+/- mice. Hum Mol Genet 19(10): 1908-1920.
- Gordon, D., Mohai, L. G. and Schwartz, S. M. (1986). Induction of polyploidy in cultures of neonatal rat aortic smooth muscle cells. Circ Res 59(6): 633-644.
- Kuang, S. Q., Kwartler, C. S., Byanova, K. L., Pham, J., Gong, L., Prakash, S. K., Huang, J., Kamm, K. E., Stull, J. T., Sweeney, H. L. and Milewicz, D. M. (2012). Rare, nonsynonymous variant in the smooth muscle-specific isoform of myosin heavy chain, MYH11, R247C, alters force generation in the aorta and phenotype of smooth muscle cells. Circ Res 110(11): 1411-1422.
- Kuang, S. Q., Medina-Martinez, O., Guo, D. C., Gong, L., Regalado, E. S., Reynolds, C. L., Boileau, C., Jondeau, G., Prakash, S. K., Kwartler, C. S., Zhu, L. Y., Peters, A. M., Duan, X. Y., Bamshad, M. J., Shendure, J., Nickerson, D. A., Santos-Cortez, R. L., Dong, X., Leal, S. M., Majesky, M. W., Swindell, E. C., Jamrich, M. and Milewicz, D. M. (2016). FOXE3 mutations predispose to thoracic aortic aneurysms and dissections. J Clin Invest 126(3): 948-961.
- Kwartler, C. S., Chen, J., Thakur, D., Li, S., Baskin, K., Wang, S., Wang, Z. V., Walker, L., Hill, J. A., Epstein, H. F., Taegtmeyer, H. and Milewicz, D. M. (2014). Overexpression of smooth muscle myosin heavy chain leads to activation of the unfolded protein response and autophagic turnover of thick filament-associated proteins in vascular smooth muscle cells. J Biol Chem 289(20): 14075-14088.
- Lemire, J. M., Covin, C. W., White, S., Giachelli, C. M. and Schwartz, S. M. (1994). Characterization of cloned aortic smooth muscle cells from young rats. Am J Pathol 144(5): 1068-1081.
- Majesky, M. W. (2007). Developmental basis of vascular smooth muscle diversity. Arterioscler Thromb Vasc Biol 27(6): 1248-1258.
- Owens, G. K. (1995). Regulation of differentiation of vascular smooth muscle cells. Physiol Rev 75(3): 487-517.
- Owens, G. K., Kumar, M. S. and Wamhoff, B. R. (2004). Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev 84(3): 767-801.
- Papke, C. L., Cao, J., Kwartler, C. S., Villamizar, C., Byanova, K. L., Lim, S. M., Sreenivasappa, H., Fischer, G., Pham, J., Rees, M., Wang, M., Chaponnier, C., Gabbiani, G., Khakoo, A. Y., Chandra, J., Trache, A., Zimmer, W. and Milewicz, D. M. (2013). Smooth muscle hyperplasia due to loss of smooth muscle α-actin is driven by activation of focal adhesion kinase, altered p53 localization and increased levels of platelet-derived growth factor receptor-β. Hum Mol Genet 22(15): 3132-27.
- Wagenseil, J. E. and Mecham, R. P. (2009). Vascular extracellular matrix and arterial mechanics. Physiol Rev 89(3): 957-989.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Kwartler, C. S., Zhou, P., Kuang, S., Duan, X., Gong, L. and Milewicz, D. M. (2016). Vascular Smooth Muscle Cell Isolation and Culture from Mouse Aorta. Bio-protocol 6(23): e2045. DOI: 10.21769/BioProtoc.2045.
Category
Developmental Biology > Cell growth and fate > Angiogenesis
Cell Biology > Cell isolation and culture > Cell isolation
Cell Biology > Cell isolation and culture > Cell differentiation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










