- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Measurement of Intracellular Calcium Concentration in Pseudomonas aeruginosa
Published: Vol 6, Iss 23, Dec 5, 2016 DOI: 10.21769/BioProtoc.2041 Views: 9316
Reviewed by: Valentine V TrotterAmit DeyAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

β-lactamase (Bla) Reporter-based System to Study Flagellar Type 3 Secretion in Salmonella
Fabienne F. V. Chevance and Kelly T. Hughes
Jun 20, 2023 1775 Views

Determination of Poly(3-hydroxybutyrate) Content in Cyanobacterium Synechocystis sp. PCC 6803 Using Acid Hydrolysis Followed by High-performance Liquid Chromatography
Janine Kaewbai-ngam [...] Tanakarn Monshupanee
Aug 20, 2023 1824 Views

An HPLC-based Assay to Study the Activity of Cyclic Diadenosine Monophosphate (C-di-AMP) Synthase DisA from Mycobacterium smegmatis
Avisek Mahapa [...] Dipankar Chatterji
Dec 20, 2024 1789 Views
Abstract
Characterization of the molecular mechanisms of calcium (Ca2+) regulation of bacterial physiology and virulence requires tools enabling measuring and monitoring the intracellular levels of free calcium (Ca2+in). Here, we describe a protocol optimized to use a recombinantly expressed Ca2+-binding protein, aequorin, for detecting Ca2+in in Pseudomonas aeruginosa. Upon binding to free Ca2+, aequorin undergoes chromophore oxidation and emits light, the log of which intensity linearly correlates with the amount of bound Ca2+, and therefore, can be used to measure the concentration of free Ca2+ available for binding. This protocol involves the introduction of the aequorin gene into P. aeruginosa, induction of apoaequorin production, reconstitution of the holoenzyme with its chromophore, and monitoring its luminescence. This protocol allows continuous measuring of Ca2+in concentration in vivo in response to various stimuli.
Keywords: Intracellular calciumBackground
Ca2+ regulates physiology and virulence of P. aeruginosa (Guragain et al., 2013; Patrauchan et al., 2005; Sarkisova et al., 2014), however, the molecular mechanisms of Ca2+ regulation are not well understood. To characterize these mechanisms, it is critically important to not only measure the concentration of Ca2+in ([Ca2+in]), but to monitor its changes in response to various stimuli. Considering that [Ca2+in] may change in response to even minute alterations in cell physiology (reviewed in [Dominguez et al., 2015]), measuring [Ca2+in] requires a tool specifically recognizing Ca2+ without significantly disturbing cells. One such tool is aequorin, a Ca2+-binding protein, which upon binding to free Ca2+, undergoes chromophore oxidation and emits light. The emitted light can be recorded as a measure of free Ca2+. Aequorin has been successfully used to monitor Ca2+in in eukaryotes (Bonora et al., 2013), as well as several bacterial species (Herbaud et al., 1998; Naseem et al., 2007; Rosch et al., 2008). Sufficient level of aequorin production and its stability within a cell enables continuous monitoring of Ca2+in (Naseem et al., 2007). Use of aequorin offers additional advantages such as targeted intracellular distribution (cytoplasm or periplasm), high dynamic range, high signal-to-noise ratio, and low Ca2+ buffering effect (Bonora et al., 2013). Alternative approaches include application of chemical indicators, such as Fura. However, due to reduced cell membrane permeability in P. aeruginosa, loading cells of this bacterium even with membrane permeable Fura acetoxymethyl (AM, ester form) is challenging and requires additional treatments, which limits physiological relevance of the measurements (not published observations). Therefore, our group pioneered the use of aequorin for measuring [Ca2+in] in P. aeruginosa (Guragain et al., 2013). The original protocol was developed for Escherichia coli (Knight et al., 1991) and further developed in (Jones et al., 1999). Here we present a modified adaptation of the protocol, successfully used to study Ca2+ homeostasis in P. aeruginosa, clinically and environmentally important organism (Guragain et al., 2013).
Materials and Reagents
- General supplies
- Strains and plasmids
- Pseudomonas aeruginosa strain PAO1 carrying pMBB66EH containing aequorin gene
- Plasmid pMBB66EH encoding aequorin gene from Aequoria victoria (courtesy: Drs. Delfina Dominguez)
- Pseudomonas aeruginosa strain PAO1 carrying pMBB66EH containing aequorin gene
- Culture medium
- Luria bertani (LB) agar (see Recipes)
- Biofilm minimal medium (BMM) (see Recipes)
- 10x basal salt solution
- Vitamin solution
- Trace metal
- 1 M MgSO4
- 10x basal salt solution
- Luria bertani (LB) agar (see Recipes)
- Chemicals and buffers
- Carbenicillin, disodium (Gold Bio, catalog number: C-103-5 )
- IPTG (Gold Bio, catalog number: I2481C )
- Calcium chloride dehydrate (CaCl2·2H2O) (Sigma-Aldrich, catalog number: C5080 )
- Live-Dead staining (Molecular Probes)
- Yeast extract (BD, Bacto, catalog number: 212750 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S7653 or VWR, catalog number: E529-500ML )
- Tryptone (BD, Bacto, catalog number: 211705 )
- Agar (BD, Bacto, catalog number: 214010 )
- Nanopure water
- Monosodium glutamate (Sigma-Aldrich, catalogue number: 1446600 )
- Glycerol (Thermo Fisher Scientific, Fisher Scientific, catalog number: G33 )
- Sodium phosphate monobasic dihydrate (NaH2PO4) (Sigma-Aldrich, catalog number: 71500 )
- Potassium phosphate dibasic (K2HPO4) (Sigma-Aldrich, catalog number: 17835 )
- Biotin (Gold Bio, catalog number: B-950-1 )
- Thiamine HCl (Gold Bio, catalog number: T-260-25 )
- HCl (Pharmco-Aaper, catalog number: 284000ACS )
- Copper(II) sulfate pentahydrate (CuSO4·5H2O) (Sigma-Aldrich, catalog number: 209198 ).
- Zinc sulfate heptahydrate (ZnSO4·7H2O) (Sigma-Aldrich, catalog number: Z0251 )
- Iron(II) sulfate heptahydrate (FeSO4·7H2O) (Sigma-Aldrich, catalog number: 215422 )
- Manganese(II) chloride tetrahydrate (MnCl2·4H2O) (Avantor Performance Materials, J.T. Baker, catalog number: 2540-04 )
- Magnesium sulfate heptahydrate (MgSO4·7H2O) (Sigma-Aldrich, catalog number: 230391 )
- HEPES (Sigma-Aldrich, catalog number: H3375 )
- MgCl2 (Sigma-Aldrich, catalog number: 230391 )
- Tergitol Type NP-40, 70% in H2O (Sigma-Aldrich, catalog number: T1135 )
- Coelenterazine (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: C2944 )
- Ethanol (Pharmco-Aaper, catalog number: AAP-111000190CSGL )
- HEPES buffer (see Recipes)
- Discharge buffer (see Recipes)
- 1 M CaCl2 solution (see Recipes)
- 6 mM CaCl2 solution for injection (see Recipes)
- Coelenterazine (see Recipes)
- Carbenicillin, disodium (Gold Bio, catalog number: C-103-5 )
Equipment
- Multichannel pipette (Finnipipette F1, Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 4661030 )
- Pipetman classic pipettes (Gilson, catalog numbers: F123600 , F123615 , F123601 , F123602 )
- 500 ml glass flasks (No specific brand is required)
- SynergyTM Mx multimode microplate reader (Biotek Instruments) with Gen5TM 2.05 PC software (BioTek Instruments)
- 37 °C incubator (Bench top incubator) (VWR, catalog number: 89409-314 )
- 37 °C shaking incubator (MaxQ 4000 table top shake incubator) (Thermo Fisher Scientific, Thermo ScientificTM, model: SHKA4000 )
- Centrifuge (Thermo Fisher Scientific, Thermo ScientificTM, model: SorvallTM RC 6 Plus Centrifuge ) with rotor type (Thermo Fisher Scientific, Thermo ScientificTM, model: F13-14x50 cy )
- Centrifuge (Eppendorf, model: 5424 )
Procedure
- Expression and reconstitution of aequorin
- Grow PAO1 strain carrying plasmid pMMB66EH encoding aequorin gene for overnight at 37 °C on LB agar plate with carbenicillin (300 μg/ml).
- Inoculate 5 ml of BMM medium with 2-3 single colonies grown on LB agar plate and incubate for 12 h at 37 °C while shaking at 200 rpm. Antibiotic was omitted hereafter to avoid its effect on bacteria.
- Transfer 1 ml of 12 h culture of OD600 0.25 (Adjust OD if needed by diluting in fresh BMM medium) to 100 ml of fresh BMM medium in 500 ml flask. Grow culture at 37 °C, 200 rpm until the mid-log phase, as determined in previous growth studies. For example, for PAO1, mid-log phase is reached at OD600 of 0.16 after 12 h of incubation.
- Once reached mid-log phase, induce cells with 1 mM IPTG (1 ml of 100 mM IPTG) and incubate for 2 h, at 37 °C, 200 rpm. At the end of induction, measure OD600 of the culture and compare with the previously recorded growth curve, to make sure that cells are still in their logarithmic phase of growth.
- Transfer cells into 250 ml ice cold centrifuge bottles. From this step and until the reconstitution step, maintain cells on ice.
- Harvest cells by centrifugation at 15,000 x g for 5 min at 4 °C. Discard supernatant and wash the cell pellet with 100 ml of ice-cold HEPES buffer. For this, first carefully resuspend cells by pipetting into of 2 ml HEPES buffer, and then add the remaining 98 ml of HEPES buffer.
- Collect cells by centrifuging at 6,000 x g for 5 min at 4 °C. Discard the supernatant.
- Resuspend collected cell pellet in 1,250 μl of HEPES buffer by pipetting and transfer 1 ml of cell suspension into a fresh microfuge tube.
- Add coelenterazine to a final concentration of 2.5 μM. For this, add 5 μl of 500 μM coelenterazine solution to 1 ml of cell suspension. Since coelenterazine is light sensitive, the procedure from this point onward must be carried out in the dark (see Notes).
- After addition of coelenterazine, incubate cell suspension at room temperature without shaking for 30 min.
Note: This step is referred to as a reconstitution step. Since coelenterazine undergoes slow oxidation in the presence of atmospheric oxygen, shaking should be strictly avoided. - Collect the cell pellet by centrifuging at 6,000 x g for 5 min at 4 °C and wash two times with 1 ml of ice-cold HEPES buffer.
- Resuspend the final collected cell pellet into 1 ml HEPES buffer by pipetting.
- Measure OD600 of thus prepared sample. Adjust the final OD600 of the cell suspension to 0.4 by adding HEPES buffer if needed.
- Grow PAO1 strain carrying plasmid pMMB66EH encoding aequorin gene for overnight at 37 °C on LB agar plate with carbenicillin (300 μg/ml).
- Measurement of luminescence
- Pipette 100 μl of cells suspension with reconstituted aequorin into 96-well luminescence (white) plate and equilibrate at room temperature for 10 min in the dark. When the effect of inhibitors or other compounds on Ca2+in to be tested, add the compounds during this step.
- Load the plate with samples into a Synergy Mx plate reader, and record luminescence for 1 min, at 5 sec interval. Use this reading to calculate a basal level of Ca2+in.
- To study the response of the intracellular Ca2+ levels to extracellular Ca2+ elevated to the millimolar concentrations commonly present in some environments including a human body, expose the cells prepared as above to the addition of 1 mM CaCl2. For this, inject 20 μl of 6 mM CaCl2 into each well by the plate reader injector. Prior to injection, prime the injector with 5 ml of 6 mM CaCl2. If the immediate effects of other compounds need to be tested, inject the latter at this step (before, after or instead of CaCl2) through a second injector, followed by luminescence measurements.
- Immediately after injection, mix the samples for 1 sec, and record luminescence for 20 min at 5 sec interval (Figure 1 A). Mixing and measurements were pre-programmed in the instrument.
- In order to estimate the remaining aequorin present in the samples, briefly take out the plate from the reader, and manually add 120 μl of discharge buffer to each sample, mix well, but quickly by pipetting, and the load back the plate into the plate reader. Read luminescence for 10 min at 5 sec interval (Figure 1B). Estimate the total aequorin by summing the luminescence detected during the entire experiment including the discharge. Use this value of total aequorin for data normalization, when calculating the concentration of Ca2+in (Figure 1C).
Note: it is important to ensure that no bubbles are formed during mixing, as bubbles will interfere with the luminescence reading.
- Pipette 100 μl of cells suspension with reconstituted aequorin into 96-well luminescence (white) plate and equilibrate at room temperature for 10 min in the dark. When the effect of inhibitors or other compounds on Ca2+in to be tested, add the compounds during this step.
Data analysis
- Calculation of free intracellular calcium concentration ([Ca2+in])
[Ca2+in] (Figure 1C) was calculated from the luminescence (Figure 1A) values using the formula: pCa = 0.612 (-log10k) + 3.745
Where,
k is a rate constant for luminescence decay (sec-1) as described in Jones et al., 1999.
[Ca2+in] at each time point was calculated as an average of at least three independent biological replicates. Even slight inconsistencies during harvesting and preparing cells may cause fluctuations in the [Ca2+in] profile. Therefore, in case of inconsistent [Ca2+in] profile, it is strongly recommended to repeat the experiment with three independent biological replicates, making sure that cells are synchronized and harvested at exactly the same point of growth. Certain mutants are particularly sensitive and produce more fluctuations in their [Ca2+in] levels. In these cases, additional replicates were added, and a [Ca2+in] profile, shared by at least 70% of biological replicates, was considered for further calculations. Replicates significantly deviating from this profile were excluded. Statistical significance was calculated by standard deviations among biological replicates (Figure 1D).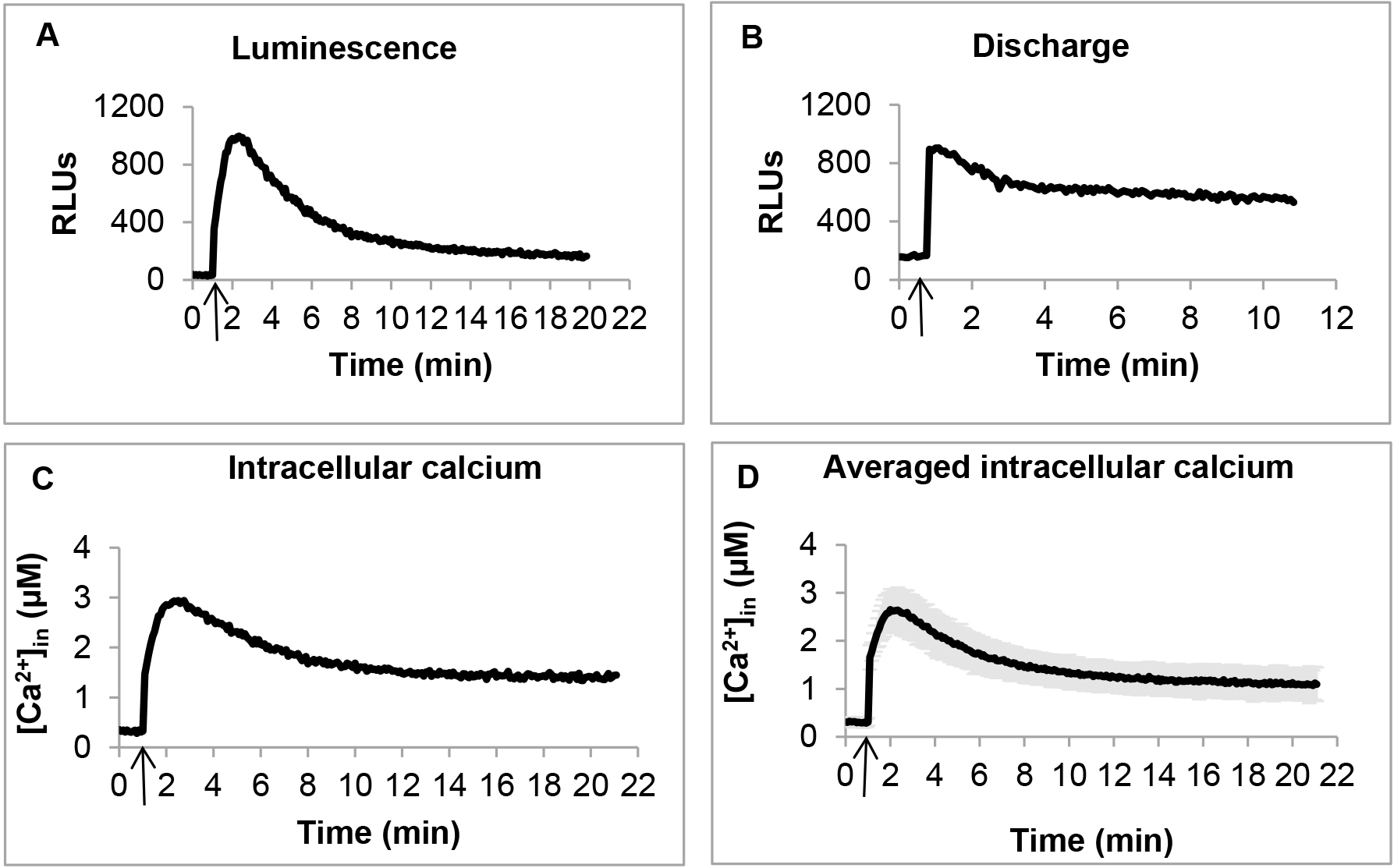
Figure 1. Measuring [Ca2+]in in PAO1 by using aequorin. A. Representative luminescence profile. Arrow indicates addition of 1 mM CaCl2. B. Representative discharge luminescence profile. Arrow indicates addition of discharge buffer (see Recipes). C. Representative calculated [Ca2+]in. Arrow indicates addition of 1 mM CaCl2. D. Averaged [Ca2+]in calculated from three independent biological replicates. Arrow indicates addition of 1 mM CaCl2.
Notes
- Two types of controls were included in the study
- Control 1 was to verify that addition of HEPES buffer alone does not affect the [Ca2+in]. For this, HEPES buffer alone was injected instead of 1 mM CaCl2 challenge, and the entire procedure was followed as described. No buffer effect was detected.
- Control 2 was to ensure that aequorin was not leaking through cell membranes. For this, 100 μl of cell suspension prepared for the measurements was incubated for 30 min in the dark at room temperature. Cells were removed by centrifugation 15,000 x g for 5 min, and the supernatant was collected, and mixed with 1 mM CaCl2. Luminescence was monitored for 1 min. We did not observe any increase in luminescence during this experiment, thus confirming that there was no aequorin leakage from the cells into the supernatant. We also verified cells viability after the procedure by Live-Dead staining (Molecular Probes).
- Luminescence profiles may vary between replicates due to possible differences in the levels of apoaequorin production. However, the calculated concentrations of Ca2+in normalized by the total available aequorin should be consistent.
- To ensure consistent [Ca2+]in measurements, it is important to induce and harvest cells at the same point of growth. During our study, we followed both OD and incubation time to determine when to harvest. Only 30 min window was allowed for cells to reach the expected density, which was estimated based on prior growth analysis.
- Aequorin is light-sensitive, and therefore all the solutions with aequorin should be kept and handled in dark tubes or tubes covered with aluminum foil. We also recommend turning off the lights in the room, only allowing either dimmed daylight or indirect light from the neighboring room. Although not tested, since aequorin absorbance maximum is at 350 nm (Shimomura and Johnson, 1969), using blue light could be safe.
- Although aequorin is reported to be stable in solution (Prendergast, 2000), we do not recommend storing cell samples with reconstituted aequorin (even on ice) for extended period of time, as this may cause fluctuations in the luminescence profiles. For this reason, only three samples were monitored at a time.
Recipes
- Luria Bertani (LB) agar
5 g yeast extract
5 g NaCl
10 g tryptone
15 g agar
Combine the ingredients in 1 L of nanopure water and autoclave - Biofilm minimal media (BMM) (Patrauchan et al., 2005)
Mix 100 ml of sterile 10x basal salt solution (Recipe 2a) to 900 ml of sterile nanopure water. Add 1 ml of vitamin solution (Recipe 2b), 200 μl of trace metals solution (Recipe 2c), and 20 μl of MgSO4 (Recipe 2d). Mix properly. - 10x basal salt solution (9.0 mM sodium glutamate, 50 mM glycerol, 0.15 mM NaH2PO4, 0.34 mM K2HPO4, 145 mM NaCl):
15 g monosodium glutamate
46 g glycerol
0.18 g sodium phosphate monobasic dehydrate (NaH2PO4)
0.78 g potassium phosphate dibasic (K2HPO4)
84.7 g sodium chloride (NaCl)
Combine the ingredients and dissolve completely in 850 ml nanopure water. Adjust the pH to 7.0. Adjust the final volume to 1,000 ml with nanopure water. Sterilize by autoclaving. - Vitamin solution
Dissolve 1 mg of biotin in 10 ml of nanopure water. Aliquot 1 ml of biotin stock solution in fresh tube and add 50 mg thiamine HCl to it and mix properly. Adjust the final volume to 100 ml with nanopure water. Filter sterilize and store at 4 °C. - Trace metal
Dilute 10 ml concentrated HCl into 70 ml of nanopure water. Add the following ingredients:
0.5 g CuSO4·5H2O
0.5 g ZnSO4·7 H2O
0.5 g FeSO4·7H2O
0.2 g MnCl2·4H2O
Dissolve completely and adjust the final volume to 100 ml with nanopure water. Filter-sterilize or autoclave. - 1 M MgSO4
24.64 g of MgSO4·7H2O was dissolved in final volume of 100 ml of nanopure water. The solution was sterilized by autoclaving. - HEPES buffer (25 mM HEPES, 125 mM NaCl, 1 mM MgCl2, pH7.5)
5.96 g HEPES
7.3 g NaCl
0.0952 MgCl2
Dissolve the ingredients in 900 ml nanopure water. Adjust the pH to 7.5 and titrate to the final volume of 1,000 ml with nanopure water. - Discharge buffer (12.5 mM CaCl2, 2% NP-40 in HEPES buffer)
To 5 ml of HEPES buffer, add 62.5 μl of 1 M CaCl2 solution and 143 μl of Tergitol. Gently mix by stirring on a magnetic stir plate to avoid foaming of NP-40 detergent - 1 M CaCl2 solution
Dissolve 36.75 g of CaCl2 dihydrate in 250 ml nanopure water. Sterilize by autoclaving - 6 mM CaCl2 solution for injection
240 μl of 1 M CaCl2 solution was added to 39.76 ml of HEPES buffer and mixed properly - Coelenterazine
Pulse-centrifuge the tube to collect the entire quantity of the reagent (250 μg coelenterazine) at the bottom. Add 1,136 μl of 95% ethanol to the tube and mix by pipetting thoroughly but quickly (to avoid light and ethanol evaporation). Resting the tube (closed) on bench for a few minutes helps dissolving. After dissolving, aliquot 50 μl of the coelenterazine solution into 0.5 ml microfuge tubes. Cover the microfuge tubes with aluminum foil and store at -20 °C
Note: Coelenterazine is highly light sensitive and should be handled in the dark.
Acknowledgments
We thank Dr. Delfina Dominguez from The University of Texas at El Paso for sharing E. coli strain carrying pMMB66EH. We thank Ian Reutlinger for transformation of P. aeruginosa PAO1 strains with pMMB66EH plasmid containing aequorin gene. This work was supported by the Grant-in-Aid from American Heart Association (Award 09BGIA2330036) and the Research Grant from OCAST (Award HR12-167).
References
- Bonora, M., Giorgi, C., Bononi, A., Marchi, S., Patergnani, S., Rimessi, A., Rizzuto, R. and Pinton, P. (2013). Subcellular calcium measurements in mammalian cells using jellyfish photoprotein aequorin-based probes. Nat Protoc 8(11): 2105-2118.
- Dominguez, D. C., Guragain, M. and Patrauchan, M. (2015). Calcium binding proteins and calcium signaling in prokaryotes. Cell Calcium 57(3): 151-165.
- Guragain, M., Lenaburg, D. L., Moore, F. S., Reutlinger, I. and Patrauchan, M. A. (2013). Calcium homeostasis in Pseudomonas aeruginosa requires multiple transporters and modulates swarming motility. Cell Calcium 54(5): 350-361.
- Herbaud, M. L., Guiseppi, A., Denizot, F., Haiech, J. and Kilhoffer, M. C. (1998). Calcium signalling in Bacillus subtilis. Biochim Biophys Acta 1448(2): 212-226.
- Jones, H. E., Holland, I. B., Baker, H. L. and Campbell, A. K. (1999). Slow changes in cytosolic free Ca2+ in Escherichia coli highlight two putative influx mechanisms in response to changes in extracellular calcium. Cell Calcium 25(3): 265-274.
- Knight, M. R., Campbell, A. K., Smith, S. M. and Trewavas, A. J. (1991). Recombinant aequorin as a probe for cytosolic free Ca2+ in Escherichia coli. FEBS Lett 282(2): 405-408.
- Naseem, R., Davies, S. R., Jones, H., Wann, K. T., Holland, I. B. and Campbell, A. K. (2007). Cytosolic Ca2+ regulates protein expression in E. coli through release from inclusion bodies. Biochem Biophys Res Commun 360(1): 33-39.
- Patrauchan, M. A., Sarkisova, S., Sauer, K. and Franklin, M. J. (2005). Calcium influences cellular and extracellular product formation during biofilm-associated growth of a marine Pseudoalteromonas sp. Microbiology 151(Pt 9): 2885-2897.
- Prendergast, F. G. (2000). Structural biology: Bioluminescence illuminated. Nature 405(6784): 291-293.
- Rosch, J. W., Sublett, J., Gao, G., Wang, Y. D. and Tuomanen, E. I. (2008). Calcium efflux is essential for bacterial survival in the eukaryotic host. Mol microbiol 70(2): 435-444.
- Sarkisova, S. A., Lotlikar, S. R., Guragain, M., Kubat, R., Cloud, J., Franklin, M. J. and Patrauchan, M. A. (2014). A Pseudomonas aeruginosa EF-hand protein, EfhP (PA4107), modulates stress responses and virulence at high calcium concentration. PLoS One 9(2): e98985.
- Shimomura, O. and Johnson, F. H. (1969). Properties of the bioluminescent protein aequorin. Biochemistry 8(10): 3991-3997.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Guragain, M., Campbell, A. K. and Patrauchan, M. A. (2016). Measurement of Intracellular Calcium Concentration in Pseudomonas aeruginosa. Bio-protocol 6(23): e2041. DOI: 10.21769/BioProtoc.2041.
Category
Microbiology > Microbial biochemistry > Other compound
Cell Biology > Cell metabolism > Other compound
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











