- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Extraction and Measurement of Abscisic Acid in a Unicellular Red Alga Cyanidioschyzon merolae
Published: Vol 6, Iss 23, Dec 5, 2016 DOI: 10.21769/BioProtoc.2033 Views: 11864
Reviewed by: Maria SinetovaSriema L. WalawageScott A M McAdam

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Evaluation of Root pH Change Through Gel Containing pH-sensitive Indicator Bromocresol Purple
Aparecida L. Silva [...] Daniel S. Moura
Apr 5, 2018 10018 Views

Quantification of Salicylic Acid (SA) and SA-glucosides in Arabidopsis thaliana
Valérie Allasia [...] Harald Keller
May 20, 2018 13141 Views

Extraction and Quantification of Plant Hormones and RNA from Pea Axillary Buds
Da Cao [...] Christine A. Beveridge
Oct 5, 2022 2968 Views
Abstract
Abscisic acid (ABA) has been known as a phytohormone of land plants, which is synthesized in response to abiotic stresses and induces various physiological responses, but is also found from eukaryotic algae. Recently, we reported that a unicellular red alga Cyanidioschyzon merolae produced ABA, which prevented cell growth and enhanced salt stress tolerance (Kobayashi et al., 2016). This report describes the detailed method for the extraction and quantification of ABA in the model red alga C. merolae.
Keywords: Abscisic acidBackground
The phytohormone ABA has been found in divergent photosynthetic eukaryotes, but the function in unicellular algae remained unclear. In a recent study, we showed that a unicellular red alga C. melorae accumulates ABA in response to salt stress by the present protocol. This is the detail of the first published protocol for the extraction and quantification of ABA from C. merolae. This protocol is optimized for C. merolae based on the land plant protocol.
Materials and Reagents
- 500 ml centrifuge bottle (Hitachi, model: S305830A )
- Membrane filters Millex-GV syringe filter unit 0.22 μm (EMD Millipore, catalog number: SLGV033RS )
- Wild type C. merolae 10D cells (National Institute for Environmental Studies, Japan)
- Abscisic acid (Sigma-Aldrich, catalog number: A4906-250UG )
- NaCl (Wako Pure Chemical Industries, catalog number: 195-15975 )
- Liquid nitrogen
- Acetic acid (Wako Pure Chemical Industries, catalog number: 017-00256 )
- Diethyl ether (Wako Pure Chemical Industries, catalog number: 052-01165 )
- Methanol (HPLC grade) (Wako Pure Chemical Industries, catalog number: 132-06471 )
- Boric acid (H3BO3) (Wako Pure Chemical Industries, catalog number: 021-15645 )
- Manganese(II) chloride tetrahydrate (MnCl2·4H2O) (Wako Pure Chemical Industries, catalog number: 133-00725 )
- Zinc sulfate heptahydrate (ZnSO4) (Wako Pure Chemical Industries, catalog number: 265-00415 )
- Sodium molybdate dehydrate (Na2MoO4·2H2O) (Wako Pure Chemical Industries, catalog number: 514-30001 )
- Copper(II) sulfate pentahydrate (CuSO4) (Wako Pure Chemical Industries, catalog number: 034-20065 )
- Cobalt(II) nitrate hexahydrate (Co[NO3]2·6H2O) (Wako Pure Chemical Industries, catalog number: 031-03752 )
- Ammonium sulfate ([NH4]2SO4) (Wako Pure Chemical Industries, catalog number: 016-03445 )
- Magnesium sulfate heptahydrate (MgSO4·7H2O) (Wako Pure Chemical Industries, catalog number: 138-00415 )
- Sulfuric acid (H2SO4) (Wako Pure Chemical Industries, catalog number: 195-04706 )
- Potassium dihydrogen phosphate (KH2PO4) (Wako Pure Chemical Industries, catalog number: 166-04255 )
- Calcium chloride (CaCl2) (Wako Pure Chemical Industries, catalog number: 036-00485 )
- Iron(III) chloride hexahydrate (FeCl3) (Wako Pure Chemical Industries, catalog number: 090-02802 )
- Na2EDTA (Wako Pure Chemical Industries, catalog number: 345-01865 )
- Polyvinylpyrrolidone K-30 (Nacalai tesque, catalog number: 28314-82 )
- 2,6-di-tert-butyl-p-cresol (Tokyo chemical industry, catalog number: D0228 )
- MA2 medium (see Recipes)
- Extraction solution (see Recipes)
Equipment
- Spectrophotometer (Beckman Coulter, model: DU730 )
- Refrigerated centrifuge (Hitachi, model: CF16RXII )
- Angle rotor (Hitachi, model: R10A3 )
- Mortar and pestle
- Vortex mixer (M & S Instruments, model: VORTEX-GENIE 2 Mixer )
- Microcentrifuge (TOMY DIGITAL BIOLOGY, model: MX150 )
- Vacuum centrifugal evaporator with low temperature trapper (TOMY DIGITAL BIOLOGY, model: CC-105 system )
- pH meter (As One, model: KR5E )
- HPLC system (Shimadzu, model: X2 HPLC system ) equipped with a photodiode array detector (PDA) and column (5 μm, 4.6 x 250 mm) (Senshu Scientific, model: ODS SP100 )
Procedure
- Extraction
- The optical density (OD) of C. merolae liquid culture is measured by spectrophotometer at 750 nm. When the OD750 reaches 10, cells are diluted to yield an OD750 of approximately 0.5 in 350 ml MA2 medium (Kobayashi et al., 2010). Grow the cells under illumination with fluorescent white light (50 μM photons m-2 s-1) at 42 °C, bubbled with air supplemented with 2% CO2. After incubation for 16 h, measure the OD750, transfer the culture to 500 ml centrifuge bottle, and collect the cells by centrifuging with angle rotor at 3,000 x g for 3 min at room temperature. Gently resuspend the pellet in 350 ml MA2 medium containing 500 mM NaCl and further cultivate for 3 h under the same condition.
- Harvest the cells (OD750 = 0.8, containing about 2 x 107 cells/ml) by centrifugation at 3,000 x g for 3 min at 4 °C, discard the medium by decantation.
- Remove the remaining medium by pipetting. Dissolve the pellet in MA2 (1-2 ml) and flash freeze in liquid nitrogen (Video 1).
- Grind the frozen cell suspension to powder by a mortar and pestle (Video 1). Video 1. Video for ABA extraction steps A3 and A4. This video supports the cell harvesting and grinding.
- Homogenize the powdered sample in 20 ml extraction solution by vortexing for 5 min and centrifuge at 10,000 x g for 15 min at 4 °C (Video 2).
- Filter the supernatant with 0.22 μm membrane filter (Video 2). Video 2. Video for ABA extraction steps A5 and A6. This video supports the cell extraction and filtration.
- Concentrate the aqueous phase by vacuum centrifugal evaporator at room temperature (collect about 4 ml aqueous phase) (Video 3).
- Measure the pH of aqueous phase by pH meter. Adjust the aqueous phase to pH 2.8 by 0.5 M acetic acid and filter with 0.22 μm membrane (Video 3).Video 3. Video for ABA extraction steps A7 and A8. This video supports the purification of extract.
- Extract the ABA from the aqueous phase by three partitions with 5 ml diethyl ether (Video 4).
- Dry up the collected ether layer by vacuum centrifugal evaporator at room temperature and dissolve the pellet in the 300 μl methanol (Video 4).Video 4. Video for ABA extraction steps A9 and A10. This video supports the continuation of purification of extract.
- The optical density (OD) of C. merolae liquid culture is measured by spectrophotometer at 750 nm. When the OD750 reaches 10, cells are diluted to yield an OD750 of approximately 0.5 in 350 ml MA2 medium (Kobayashi et al., 2010). Grow the cells under illumination with fluorescent white light (50 μM photons m-2 s-1) at 42 °C, bubbled with air supplemented with 2% CO2. After incubation for 16 h, measure the OD750, transfer the culture to 500 ml centrifuge bottle, and collect the cells by centrifuging with angle rotor at 3,000 x g for 3 min at room temperature. Gently resuspend the pellet in 350 ml MA2 medium containing 500 mM NaCl and further cultivate for 3 h under the same condition.
- HPLC analysis
- Chromatography is conducted on an HPLC system equipped with a PDA using the full visible spectrum with monitoring employed at 254 nm. Column is a tandem jointed ODS SP100, 250 x 4.6 mm, S = 5 µm.
- Perform isocratic separation by 50% methanol. Flow rate is 0.5 ml/min at 40 °C, and apply the samples in 10-50 μl. The peak of ABA was detected at retention time 7.1 min (Figure 1).
- Standard calibration curves were generated at 254 nm with ABA reference.
- Standard ABAs of concentration of 0, 10, 50, 100, 200, 500, 1000 pmol/μl are prepared and applied to HPLC in 10 μl. HPLC analysis should be performed in triplicates.
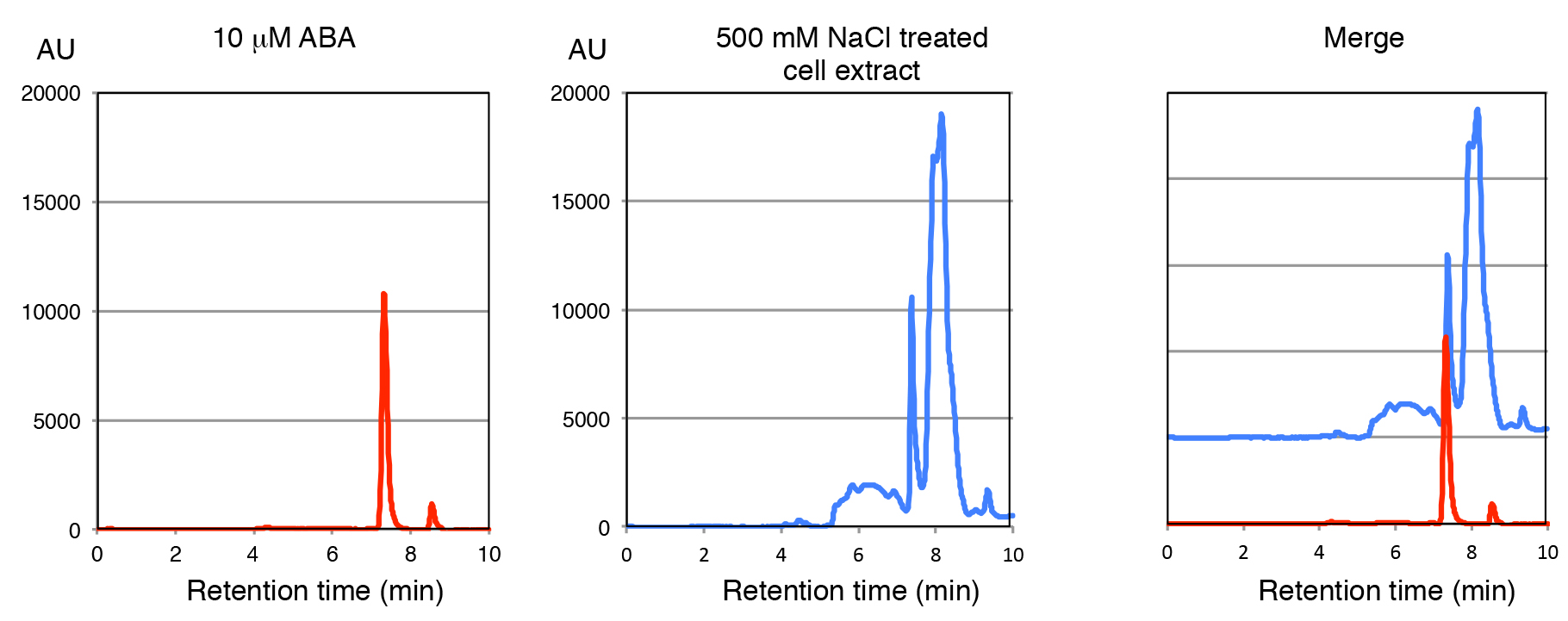
Figure 1. HPLC separation of the C. merolae extract with ABA standard reference. Cells were treated with 500 mM NaCl for 3 h. The ABA standard reference and 500 mM NaCl treated cell extract were separated by HPLC with PDA. The absorption spectra of 0.2 mM ABA standard (left panel), 500 mM NaCl treated cell extract (center panel) and merged (right panel) are shown.
Data analysis
- The retention time of the single peak areas is recorded and calculate the average of each peak.
- Create a calibration curve using the average of each peak. Calculate the ABA amount using a specific standard calibration curve (An example is shown in Figure 2).
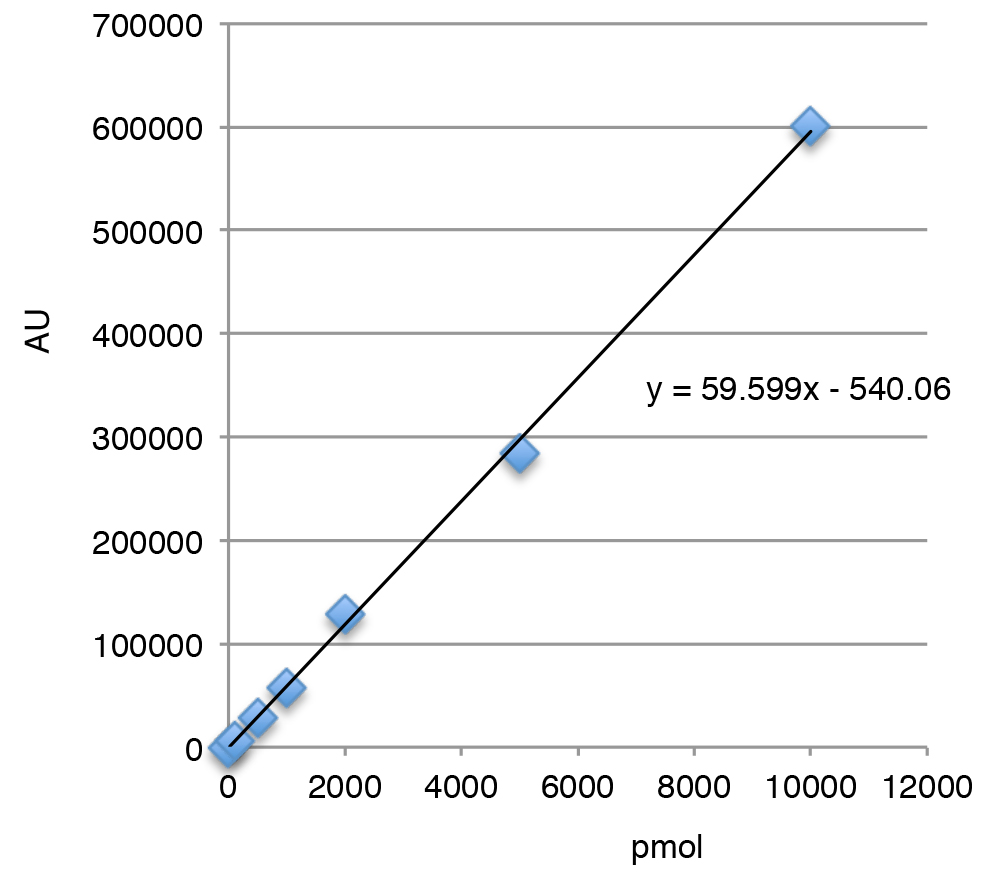
Figure 2. ABA standard curve. Standard curve was made based on peak value in HPLC analysis. Example: 300 μl extract was obtained starting from salt stressed cell culture (350 ml, OD750 = 0.8), and 10 μl of the 300 μl was subjected to HPLC analysis, which resulted in the read of 133.533 AU. Based on the calibration curve, the total amount of ABA contained in the starting material was calculated as 68487.5 pmole.
Notes
ABA extraction and HPLC method was modified from Kojima method (Kojima et al., 1995). ABA is detected from C. merolae cells only under the salt stressed condition.
Recipes
- MA2 medium
Mix solutions I to III and mess up to 1 L. Sterilize by autoclaving. Solution IV should be sterilized by filtration through a 0.22-micron filter. Add solution IV to the mix after autoclave.
Solution I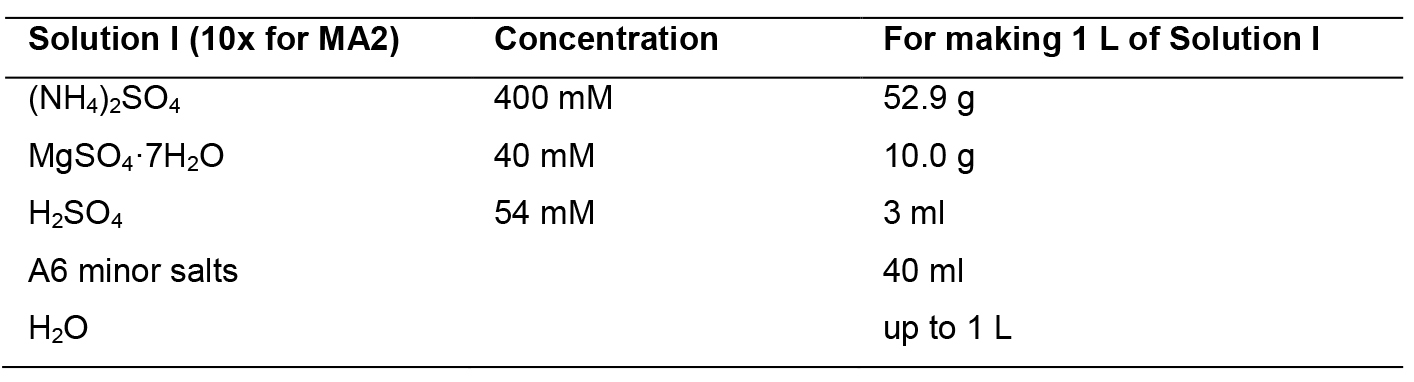
Solution II
Solution III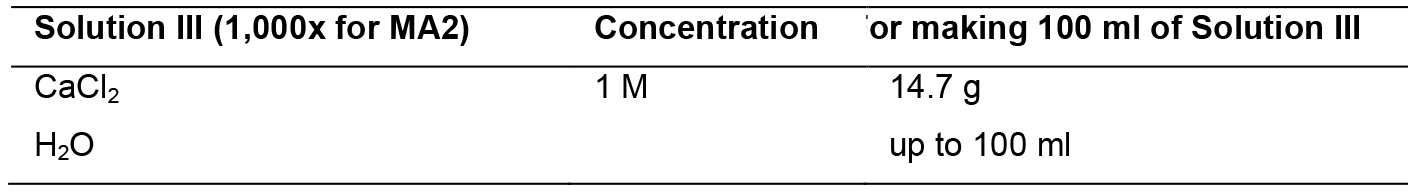
Solution IV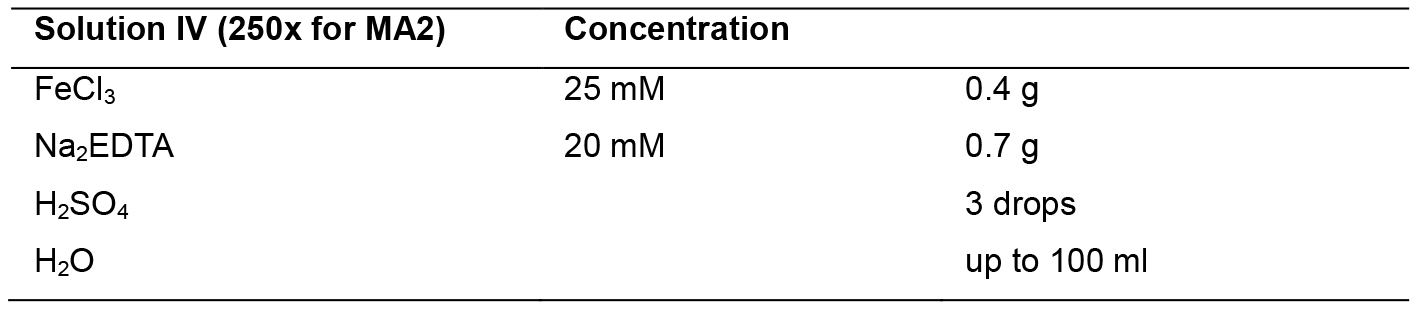
A6 minor salts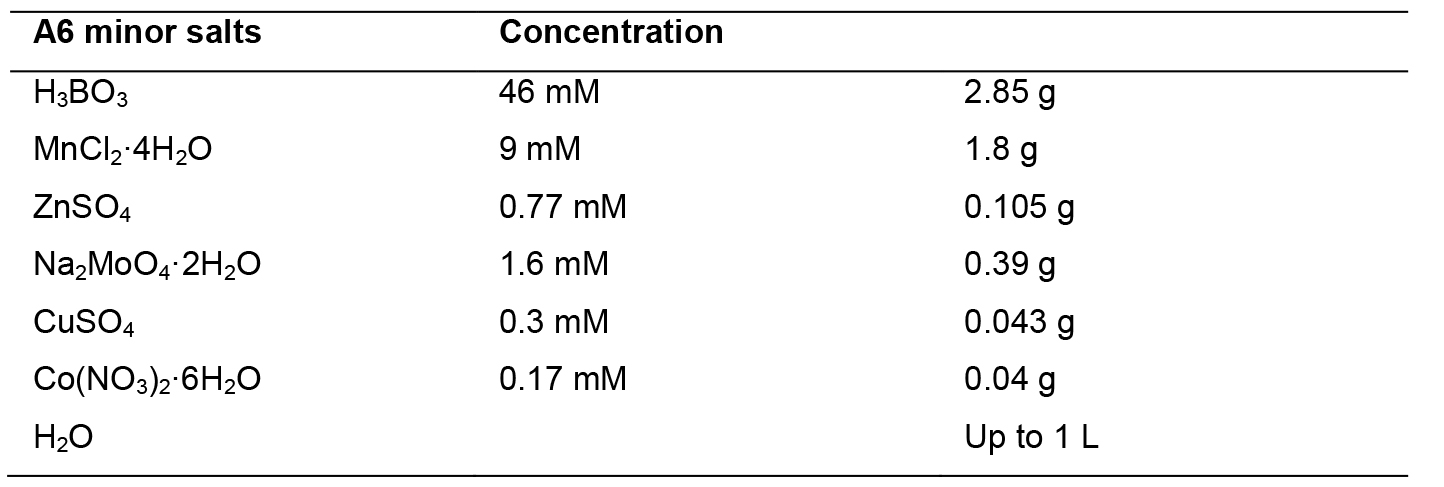
- Extraction solution
0.6 g/L polyvinylpyrrolidone
0.22 g/L 2,6-di-tert-butyl-p-cresol
Dissolve in 80% (v/v) methanol
Acknowledgments
This protocol was adapted from Kobayashi et al. (2016). The authors thank Dr. Tadao Asami for technical help in ABA detection. This study was supported by MEXT/JSPS KAKENHI (Grant numbers: 21370015, 23120505, 2424806, 15K14539 to K.T., 13274350, 15621958 to Y.K.)
References
- Kobayashi, Y., Ando, H., Hanaoka, M. and Tanaka, K. (2016). Abscisic acid participates in the control of cell-cycle initiation through heme homeostasis in the unicellular red alga Cyanidioschyzon merolae. Plant Cell Physiol 57(5), 953-960.
- Kobayashi, Y., Ohnuma, M., Kuroiwa, T., Tanaka, K. and Hanaoka, M. (2010). The basics of cultivation and molecular genetic analysis of the unicellular red alga Cyanidioschyzon merolae. Endocytobiosis Cell Res 20: 53-61.
- Kojima, K., Yamada, Y. and Yamamoto, M. (1995). Effects of abscisic acid injection on sugar and organic acid contents of citrus fruit. J Japan Soc Hort Sci 64(1): 17-21.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Kobayashi, Y. and Tanaka, K. (2016). Extraction and Measurement of Abscisic Acid in a Unicellular Red Alga Cyanidioschyzon merolae. Bio-protocol 6(23): e2033. DOI: 10.21769/BioProtoc.2033.
Category
Plant Science > Plant biochemistry > Plant hormone
Biochemistry > Other compound > Plant hormone
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












