- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Uptake Assay for Radiolabeled Peptides in Yeast
Published: Vol 6, Iss 22, Nov 20, 2016 DOI: 10.21769/BioProtoc.2026 Views: 9207
Reviewed by: Yanjie LiChong HeMelike Çağlayan

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Live Cell Measurement of the Intracellular pH of Yeast by Flow Cytometry Using a Genetically-Encoded Fluorescent Reporter
Catherine G. Triandafillou and D. Allan Drummond
Jun 20, 2020 4827 Views

Polyamine Transport Assay Using Reconstituted Yeast Membranes
Sarah van Veen [...] Peter Vangheluwe
Jan 20, 2021 4505 Views

Single Cell Analysis and Sorting of Aspergillus fumigatus by Flow Cytometry
Gareth Howell and Robert-Jan Bleichrodt
Apr 20, 2021 5296 Views
Abstract
We describe an assay for measuring the uptake of radioactive peptides into the yeast Saccharomyces cerevisiae. The methods presented here can be adapted to measure a variety of substrates transported into any bacterial or fungal cell via specific carrier-mediated systems.
Background
Di/tripeptides and larger oligopeptides are sources of amino acids for protein synthesis, or may serve as carbon and nitrogen precursors for energy production and biosynthesis of metabolites in all organisms. Uptake of di/tripeptides and oligopeptides across the cell membrane is facilitated by peptide transporters. Measuring the accumulation of radiolabeled peptide provides an experimental approach to determine uptake across a specific peptide transport protein in a variety of cell types, including the yeast Saccharomyces cerevisiae. Appropriate experimental design allows for the determination of kinetic parameters such as the affinity and capacity of system under investigation. Dipeptides, such as radiolabeled Leu-Leu, can be used to measure transport across the di/tripeptide transporter Ptr2p of S. cerevisiae. We use the transport of dipeptides across Ptr2p in yeast as an example.
Materials and Reagents
- Culture tube, up to 17 x 100 mm (Thermo Fisher Scientific, Fisher Scientific, catalog number: 14-956-6B )
- 1.5 ml microfuge tubes
- HAWP membrane filter (EMD Millipore, catalog number: HAWP02400 )
- Saccharomyces cerevisiae (S. cerevisiae): such as W303 strain (MATa or MATα leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15) (ATCC, catalog number: 208352 )
Note: Other strains can be used, although the growth medium will have to be adjusted if specific auxotrophic requirements are needed. - Proline (Sigma-Aldrich, catalog number: P5607 )
- Yeast nitrogen base without (NH4)2SO4 and amino acids (BD, catalog number: 233510 )
- D-glucose (Sigma-Aldrich, catalog number: G7021 )
- Uracil (Sigma-Aldrich, catalog number: U1128 )
- Adenine hemisulfate salt (Sigma-Aldrich, catalog number: A9126 )
- Histidine-HCl (Sigma-Aldrich, catalog number: H-8000 )
- Leucine (Sigma-Aldrich, catalog number: L8000 )
- Tryptophan (Sigma-Aldrich, catalog number: T0254 )
- Radiolabeled dipeptide [3H]Leu-Leu (Custom synthesized)
- Leu-Leu (Sigma-Aldrich, catalog number: L-2752 )
- Minimal Proline (MP) broth (see Recipes)
- MP+ broth (see Recipes)
- 20% glucose (see Recipes)
- 2% glucose (see Recipes)
- 2x uptake medium (see Recipes)
Equipment
- 250 ml flask
- Vortex mixer (VWR, catalog number: 97043-562 )
- Water bath (VWR, catalog number: 89501-476 )
- Microscope
- Incubator (set up at 30 °C) containing a rotator mixer (Thermo Fisher ScientificTM, Thermo Scientific model: 1640Q )
- Shaker incubator, 30 °C (Eppendorf, New Brunswick Scientific, model: C-25 )
- Hemocytometer
- Dry bath, 30 °C with blocks to hold microfuge 1.5 ml tubes (Thermo Fisher Scientific, catalog number: 88870002 )
- Manifold vacuum filtration cell harvester (EMD Millipore, catalog number: xx2702550 )
- Filter forceps (EMD Millipore, catalog number: XX6200006P )
- Liquid Scintillation vial, 6 ml Omni-vial with cap (Wheaton, catalog number: 225414 )
- Liquid Scintillation Cocktail (MP Biomedical CytoScint, catalog number: 882453 )
- Liquid Scintillation Counter, TRI-CARB 2000TR (PerkinElmer, catalog number: B291000 )
Procedure
- Preparation of a working cell stock
Yeast cells are grown in a culture tube containing 5 ml of minimal medium supplemented with proline as the nitrogen (N) source (MP) and compounds to satisfy the auxotrophic requirements (MP+ broth, see Recipes below) at 30 °C overnight in a rotator until a turbid culture is obtained. The minimal medium described in this protocol is designed for the W303 strain. Proline is added as a poor N source in order to induce the synthesis of the peptide uptake system of S. cerevisiae. Other minimal media can be used for strains of S. cerevisiae with different auxotrophic requirements. - 1 ml of the above overnight culture is seeded into a 250 ml flask containing 50 ml of fresh MP+ broth and grown to log phase, corresponding to a density of approximately 5 x 106 cells/ml, taking about 4 h.
- Log phase cells are harvested by centrifugation (1,000 x g) and washed three times with a solution of 2% glucose to remove culture medium residue and maintain the cells in an energized state.
- Adjustment of the cell number
The cell pellet is resuspended in 0.5 ml 2% glucose and the suspended cells are counted on a hemocytometer. Based on the hemocytometer results, a cell suspension is prepared at a final concentration of 1 x 108 cells/ml in 2% glucose. - Setting-up the cell uptake
The 2x uptake medium is warmed to 30 °C. 60 μl of cell suspension is dispensed into each of ten, 1.5 ml microfuge tubes. The tubes are transferred into a 30 °C dry bath block and incubated for 5 min. To begin the assay, 60 μl of pre-warmed, 2x uptake medium are added to the pre-warmed 60 μl yeast cell suspensions and the suspension carefully mixed by pipetting. The tubes are incubated for various time intervals. Generally, times should range from a minimum of 10 sec to a maximum of 10 min. For each uptake time, triplicate determinations are made. To control for non-specific association of the radiolabeled peptide with the cells, a negative control in which no peptide should be transported must be completed. This can be done by repeating the entire uptake procedure at 0 °C, when cells are not metabolically active. Alternatively, completing the uptake assay at 30 °C using an isogenic strain known to be lacking the transporter of interest can also serve as a negative control. - Stopping the uptake
Upon completion of the uptake interval, cells are harvested and washed by filtration on any manifold system such as the Manifold Vacuum Filtration Cell Harvester (see Equipment list). A membrane filter pre-wet in water (one filter is needed for each of the ten samples) is placed onto the filtration apparatus, the apparatus is assembled and the vacuum is turned on. To stop the uptake, 100 μl of the uptake/cell suspension mixtures are pipetted onto the center of the filter. Each filter is then washed 4 times with 1 ml of ice-cold water.
Notes:- The HAWP membrane filters needed to complete the assay are placed into a Petri dish with water to pre-wet. Filters are removed as needed and placed onto the filtration apparatus just prior to stopping the uptake assay.
- It is important to avoid loading the cell suspension near the rim of the filter as this could result in loss of cells if the filtration apparatus does not seal tightly around the filter.
- The HAWP membrane filters needed to complete the assay are placed into a Petri dish with water to pre-wet. Filters are removed as needed and placed onto the filtration apparatus just prior to stopping the uptake assay.
- Radioactivity measurement
Turn off the vacuum to the filtration device and disassemble to access the filters. Remove the filters with the retained yeast cells using filter forceps and place each into a separate liquid scintillation vial. One milliliter of scintillation fluid is added to each vial, which is capped and then shaken by hand to ensure that the filter is completely submerged in the cocktail. The vials are allowed to sit at room temperature for a minimum of one hour to allow for equilibration with the scintillation fluid and radioactivity is measured using a Liquid Scintillation Counter, per manufacturer’s instructions.
Data analysis
The uptake of the radioactive substrate is plotted in terms of nmol of substrate/109 cells vs. time. For each uptake time, triplicate determinations are made, averaged, and the mean ± standard deviation are plotted. To determine the specific uptake, subtract the negative control values from the total uptake. A representative graph for the uptake of radiolabeled Leu-Leu is presented below (Figure 1).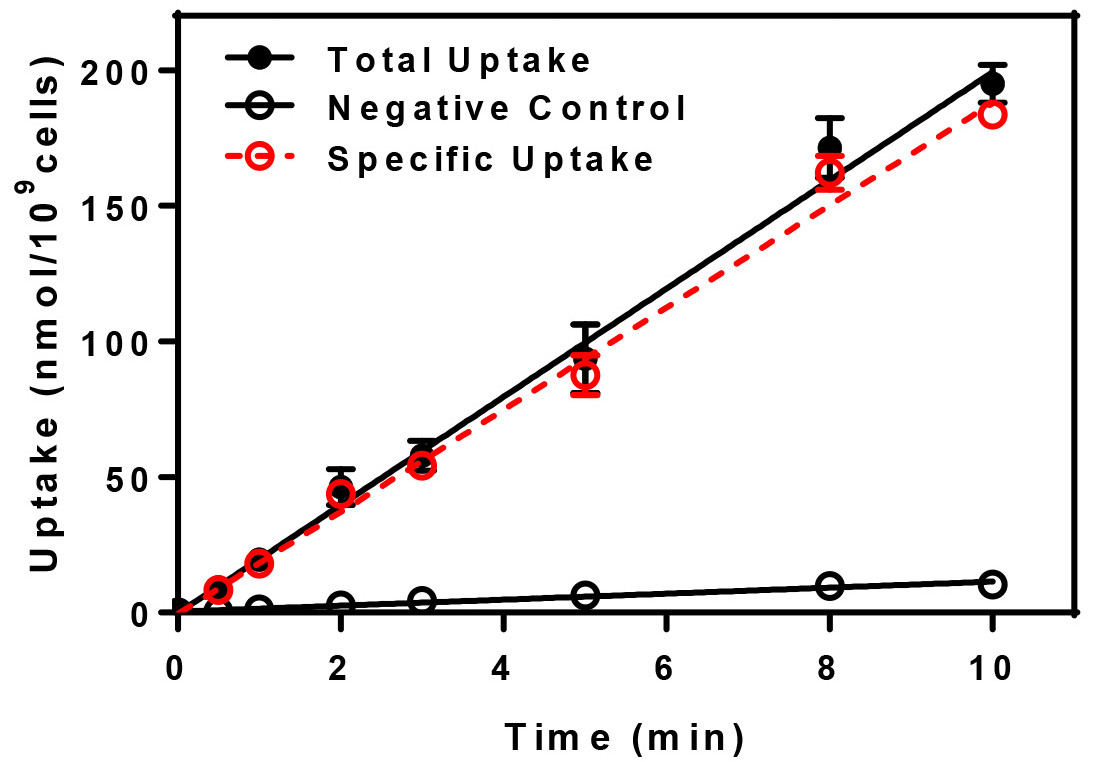
Figure 1. Uptake vs. time. Uptake of radiolabeled dileucine was determined for yeast cells at 30 °C (Total Uptake) and at 0 °C (Negative Control) and normalized to reflect uptake in nmol per 109 yeast cells. The specific uptake, indicated in red, is determined by subtracting the negative control from the total.
Notes
- The experiment described above measures accumulation, or uptake, of the substrate into the cell. If kinetic measurements of transport are to be made, then it is essential that the intracellular fate of the substrate under study is understood. For example, should the peptide be rapidly hydrolyzed during transport or immediately upon entering the cytoplasm, the determination of kinetics by the method described above is not valid. It is also essential to determine that the substrate is not metabolized extracellularly before transport as well. If extracellular hydrolysis occurs, then that substrate would not be suitable for uptake measurements because uptake of the radioactivity would be measuring amino acid uptake. Therefore, extraction and analysis of the extracellular medium and the intracellular contents after transport measurement to determine the physical state of the substrate is essential for kinetic transport studies, whereas if only assessing ‘uptake’ is the goal of the studies, then such determination is not necessary.
- This experiment may be repeated with various amounts of non-metabolized radioactive substrate to determine the kinetics of uptake by way of Michaelis-Menten type analysis. Uptake is measured at a time in the linear phase of accumulation as determined by measurement of uptake vs. time (Figure 1) at various substrate concentrations. For each uptake point, triplicate determinations were made, averaged, and the mean ± standard deviation are plotted. The plot of substrate concentration (S) vs. uptake (V) will yield the apparent Km and the apparent Vmax of the transport system under study. Vmax represents the maximum rate achieved by the system and the Km is the substrate concentration at which the uptake rate is half of the maximum. A representative analysis of transport kinetics is provided below (Figure 2).
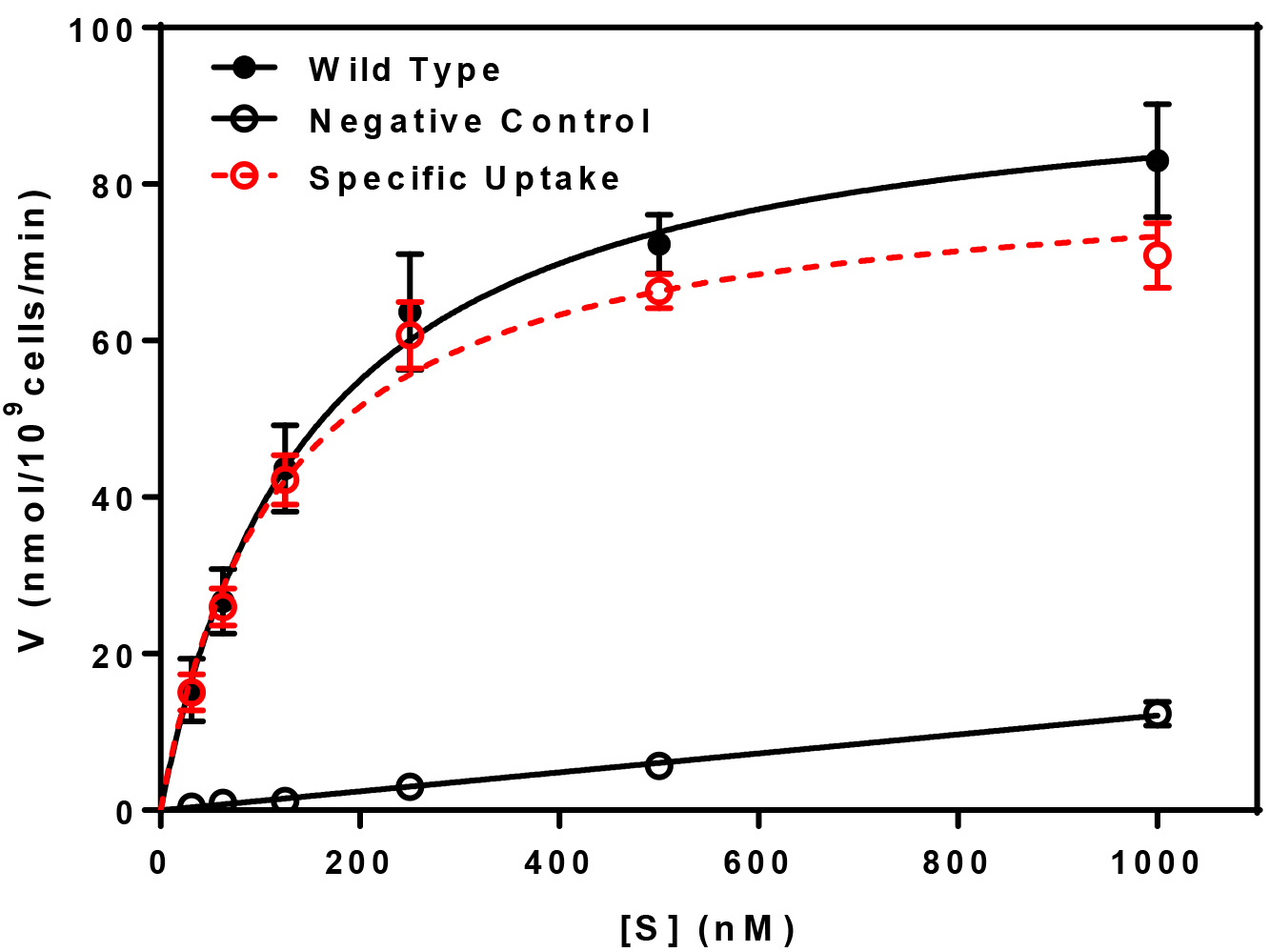
Figure 2. Kinetics of peptide transport. Uptake (V) was determined as a function of [3H]Leu-Leu concentration (S) at 30 °C (Total Uptake) and at 0 °C (Negative Control). Values were normalized to reflect uptake in nmol per 109 yeast cells. The specific uptake, indicated in red, is determined by subtracting the negative control from the total. Data were fitted by non-linear regression to determine the Km (118 ± 18 nM) and the predicted Vmax (82 ± 4 nmol/109 cells/min) values for the specific uptake component.
Recipes
- Minimal Proline (MP) broth (1L)
20 g glucose
1.7 g yeast nitrogen base (YNB) without (NH4)2SO4 and amino acids
1 g of proline as a nitrogen source.
The glucose is dissolved in 900 ml water and autoclave-sterilized
Once cooled, 100 ml of a filter sterilized 10x YNB-proline stock (17 g YNB and 10 g proline in 100 ml water) is added
Note: Proline is used as a nitrogen source to induce the synthesis of the yeast peptide transport system. - MP+ broth
100 ml of MP broth
1.0 ml of uracil (2 mg/ml in double distilled water, filter-sterilized)
1.0 ml adenine sulfate (2 mg/ml 0.1 N HCl, filter-sterilized)
0.2 ml of histidine (10 mg/ml, filter-sterilized)
0.3 ml of leucine (10 mg/ml, filter-sterilized)
0.2 ml of tryptophan (10 mg/ml 0.1 N HCl, filter-sterilized)
Note: The above amino acids and nucleobases are added to satisfy the auxotrophic requirement of S. cerevisiae W303. The specific growth conditions may be modified for the yeast strain or toxic compound to be assayed. - 20% glucose (100 ml)
Per 100 ml contains 20 g glucose
Dissolve the glucose in double distilled water
Adjust the final volume to 100 ml and autoclave or filter-sterilize the solution - 2% glucose
Dilute the 20% glucose solution 1:10 in water - 2x uptake medium
This will be diluted 1:1 with the cell mixture to begin the uptake assay
The 2x medium consists of 2% glucose and 20 mM sodium citrate-potassium phosphate (pH 5.5)
This is supplemented with radiolabeled peptide [2 µCi/ml (14C) Leu-Leu] and non-radiolabeled peptide (150 µM Leu-Leu)
Note: The amount of substrate (radiolabeled and non-radiolabeled peptide) included in the 2x uptake medium will vary based on the nature of the experiment (Uptake vs. Time or Uptake vs. Substrate Concentration) and must be determined empirically for each cell type. The values given above are a starting point.
Acknowledgments
This protocol was adapted from our previous studies (Cai et al., 2007; Cai et al., 2006). This work was supported by grants from the National Institute of General Medical Sciences GM-22087 and GM-46520.
References
- Cai, H., Kauffman, S., Naider, F. and Becker, J. M. (2006). Genomewide screen reveals a wide regulatory network for di/tripeptide utilization in Saccharomyces cerevisiae. Genetics 172(3): 1459-1476.
- Cai, H., Hauser, M., Naider, F. and Becker, J. M. (2007). Differential regulation and substrate preferences in two peptide transporters of Saccharomyces cerevisiae. Eukaryot Cell 6(10): 1805-1813.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Hauser, M., Cai, H., Naider, F. and Becker, J. M. (2016). Uptake Assay for Radiolabeled Peptides in Yeast. Bio-protocol 6(22): e2026. DOI: 10.21769/BioProtoc.2026.
Category
Microbiology > Microbial cell biology > Cell-based analysis
Cell Biology > Cell-based analysis > Transport
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link