- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Halo Assay for Toxic Peptides and Other Compounds in Microorganisms
Published: Vol 6, Iss 22, Nov 20, 2016 DOI: 10.21769/BioProtoc.2025 Views: 9275
Reviewed by: Yanjie LiChong HeAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Live Cell Measurement of the Intracellular pH of Yeast by Flow Cytometry Using a Genetically-Encoded Fluorescent Reporter
Catherine G. Triandafillou and D. Allan Drummond
Jun 20, 2020 4826 Views

Polyamine Transport Assay Using Reconstituted Yeast Membranes
Sarah van Veen [...] Peter Vangheluwe
Jan 20, 2021 4504 Views

Single Cell Analysis and Sorting of Aspergillus fumigatus by Flow Cytometry
Gareth Howell and Robert-Jan Bleichrodt
Apr 20, 2021 5295 Views
Abstract
We describe an assay for determination of toxicity in S. cerevisiae involving spotting of a toxic peptide on a lawn of yeast cells. This assay may be generalized to determine toxicity of a variety of compounds by substituting a putative toxic compound in place of the peptide. The general protocol may also be used to determine toxicity of any small compound toward another microorganism by replacing S. cerevisiae with the target microbe and modifying growth conditions accordingly.
Background
Di-/tripeptides are one of the major sources of nitrogen, carbon, and amino acids for all organisms. Synthetic peptides containing a toxic amino acid residue provide an experimental approach to measure peptide transport and/or utilization in Saccharomyces cerevisiae. Hydrolysis of internalized peptides by intracellular peptidases or proteases releases the toxic residue leading to an easily detectable zone (halo) of growth arrest on a lawn of cells plated in a Petri plate. For example, upon intracellular hydrolysis the toxic peptide Ala-Eth releases ethionine (Eth), a methionine antagonist which interferes with the incorporation of amino acids into proteins and with the normal methylation of DNA and other methylation pathways, thereby leading to cell death. When spotted onto a lawn of yeast cells, the transported dipeptide Ala-Eth will inhibit growth, and a clear ‘halo’ will form in the lawn of cells around the region where the Eth-containing toxic peptide is spotted (Figure 1A). The assay described here for determination of peptide toxicity in S. cerevisiae may be generalized as follows: (1) it may be modified to determine toxicity of any substrate by simply using a putative toxic compound in place of a peptide containing a toxic amino acid, or (2) it may be modified to determine toxicity of a substrate toward any microorganism by replacing S. cerevisiae in the assay with the target organism. It is a simple, inexpensive and relatively rapid method for determining substrate toxicities as modified for the specific organism and toxic moiety assayed.
Materials and Reagents
- 16 x 125 mm glass screw cap tubes (Thermo Fisher Scientific, Fisher Scientific, catalog number: 14-959-25C )
- 6-mm sterile blank paper discs (BD, catalog number: 231039 )
- Culture tube (Thermo Fisher Scientific, Fisher Scientific, catalog number: 14-956-6B )
- Sterile Petri plate (Thermo Fisher Scientific, Fisher Scientific, catalog number: FB0875713 )
- Saccharomyces cerevisiae (S. cerevisiae): such as W303 strain (MATa or MATα leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11, 15) (ATCC, catalog number: 208352 )
Note: Other strains can be used, although the growth medium will have to be adjusted to meet specific auxotrophic requirements. - Toxic peptides, such as Ala-Eth or Leu-Eth (Custom synthesized)
- Proline (Sigma-Aldrich, catalog number: P5607 )
- Agar (Sigma-Aldrich, catalog number: A1296 )
- Yeast nitrogen base without (NH4)2SO4 and amino acids (BD, DifcoTM, catalog number: 233510 )
- Dextrose (Sigma-Aldrich, catalog number: G7021 )
- Uracil (Sigma-Aldrich, catalog number: U1128 )
- Adenine hemisulfate salt (Sigma-Aldrich, catalog number: A9126 )
- Histidine-HCl (Sigma-Aldrich, catalog number: H-8000 )
- Leucine (Sigma-Aldrich, catalog number: L8000 )
- Tryptophan (Sigma-Aldrich, catalog number: T0254 )
- Noble agar (Sigma-Aldrich, catalog number: A5431 )
- Minimal proline (MP) broth (see Recipes)
- MP+ broth (see Recipes)
- MP+ growth plate (see Recipes)
- 1.1% Noble agar blanks (see Recipes)
Equipment
- Ruler
- Filter forceps (EMD Millipore, catalog number: XX6200006P )
- Vortex mixer (VWR, catalog number: 97043-562 )
- General purpose water bath (VWR, catalog number: 89501-476 )
- Microscope
Note: Any standard lab microscope can be used. - Incubator (set up at 30 °C) containing a rotator mixer (Thermo Fisher Scientific, Thermo ScientificTM, model: 1640Q )
- Hemocytometer (Thermo Fisher Scientific, Fisher Scientific, catalog number: 0267151B )
- Tabletop centrifuge (Eppendorf, model: 5810R )
Software
- ImageJ (www.imagej.net)
Procedure
- Prepare a working cell stock
Yeast cells taken from a freshly steaked MP+ growth plate (see Recipes) are grown in a culture tube (see Materials and Reagents) containing 5 ml MP+ broth (see Recipes) at 30 °C overnight in a rotator at 100 rpm until a turbid culture is obtained. The minimal medium described in this protocol is designed for the W303 strain. Proline is added as a poor N source in order to induce the synthesis of the peptide uptake system of S. cerevisiae. Other minimal media can be used for strains of S. cerevisiae with different auxotrophic requirements. - The overnight culture is harvested by low-speed centrifugation in the table top centrifuge (1,000 x g) for 5 min at room temperature and washed three times with 5.0 ml of sterile, distilled water under the same centrifugation conditions to remove the culture medium.
- Adjusting the cell number
The cell pellet is resuspended in 0.5 ml of water, and the suspended cells are counted on a hemocytometer. Based on the hemocytometer results a cell suspension is prepared at a final concentration of 5 x 106 cells/ml in water. 1 ml of cells will be needed for each test plate. - Pouring the top agar
1 ml of the 5 x 106 cells/ml suspension is added to a tube of molten top agar (3.0 ml Noble agar blank) previously equilibrated to 55 °C, for a top-agar overlay. After briefly mixing with a vortex, the cell/top agar mixture is immediately poured onto a Petri plate containing 25 ml of MP+ growth plate (see Recipes). The top agar is poured quickly and the plate is gently swirled to ensure that the top agar mixture is evenly spread across the surface of the plate.
Notes:- 1.1% Noble agar will solidify quickly at room temperature. The culture tubes holding the Noble agar need to be placed in a boiling water bath to melt the agar, then placed in a 55 °C water bath or dry block to maintain in the liquid state.
- Pouring top agar onto the MP+ growth plate should be done as quickly as possible to prevent the Noble agar mixture from solidifying before spreading. Pre-warming the MP+ growth plate in a 30 °C incubator is helpful while developing the skills for a successful pour. It is very important to spread the top agar evenly on the surface of the MP+ growth plate to generate a uniform lawn. If a bubble forms after pouring, it can be popped by touching it with a sterile pipette tip. No Bunsen burner or hood is needed if poured carefully.
- 1.1% Noble agar will solidify quickly at room temperature. The culture tubes holding the Noble agar need to be placed in a boiling water bath to melt the agar, then placed in a 55 °C water bath or dry block to maintain in the liquid state.
- Loading toxic peptides
Sterile paper disks are placed into a clean, sterile Petri plate. Serial dilutions of the peptide to be tested are prepared and 10 µl of each solution is placed onto a sterile paper disk. For statistical accuracy, this should be done in triplicate. Once the peptide solution has been absorbed into the disk, sterile filter forceps are used to transfer the disk onto the top agar lawn; one disk should be placed in the center of the plate, with the others spaced evenly around the edge (Figure 1A). Different amounts of the toxic peptide will result in a zone of growth arrest or ‘halo’ around the disk. The diameter of the halo is proportional to the amount of toxic compound added.
Notes:- Since the surface of the plate is moist, gently placing the disk with forceps onto the surface of the top agar is sufficient for it to adhere to the plate. Pressing the disk onto the top agar should be avoided as it may deform the surface of the top agar.
- The toxic peptide on the disk will diffuse from the disk through the top agar lawn of cells. The development of the halo is dependent on the concentration of the toxic peptide surrounding the disk. It is essential that the toxic peptide is absorbed uniformly into the disk to ensure that the resulting halo is symmetric. To ensure this, slowly add the solution of the toxic peptide to the center of the paper disk.
- Different toxic substrates may diffuse to different extents in the agar. The amount of toxic substrate added to a disk must be adjusted for that specific substrate.
- Since the surface of the plate is moist, gently placing the disk with forceps onto the surface of the top agar is sufficient for it to adhere to the plate. Pressing the disk onto the top agar should be avoided as it may deform the surface of the top agar.
- Incubation
Once the disks have been placed on the top agar lawn, place the plate upside down in a 30 °C incubator for 1-2 days. Plates are incubated in this inverted position to prevent condensation onto the lawn and disks, which will disrupt the halo assay.
Data analysis
- Halo size determination
The halo diameter is measured using a ruler and recorded to the nearest 0.5 mm. Alternately, the plates can be imaged and halo diameter and/or area can be determined using ImageJ (www.imagej.net) or similar software. The measurements are averaged for the three replicates and quantitative evaluation of the toxicity can be done by plotting the size of the halo against the amount of toxic moiety added to a disk. A log plot of amount of toxic peptide added versus halo size normally yields linear results within the range of concentrations measured (Figure 1B). - Evaluation of results
A clear zone of inhibition after a 24-48 h incubation may indicate either a static (inhibits cell growth) or cidal (cells are killed) effect. If the clear halo persists after continuing the incubation for another 2-4 days, then this would indicate a cidal effect. If the halo fills in uniformly after the additional incubation, the toxic moiety was probably static. In some instances, a clone arising in the halo zone might indicate growth of a resistant mutant that arose spontaneously or was preexistent in the cells plated. This resistant clone might be a valuable resource for further study.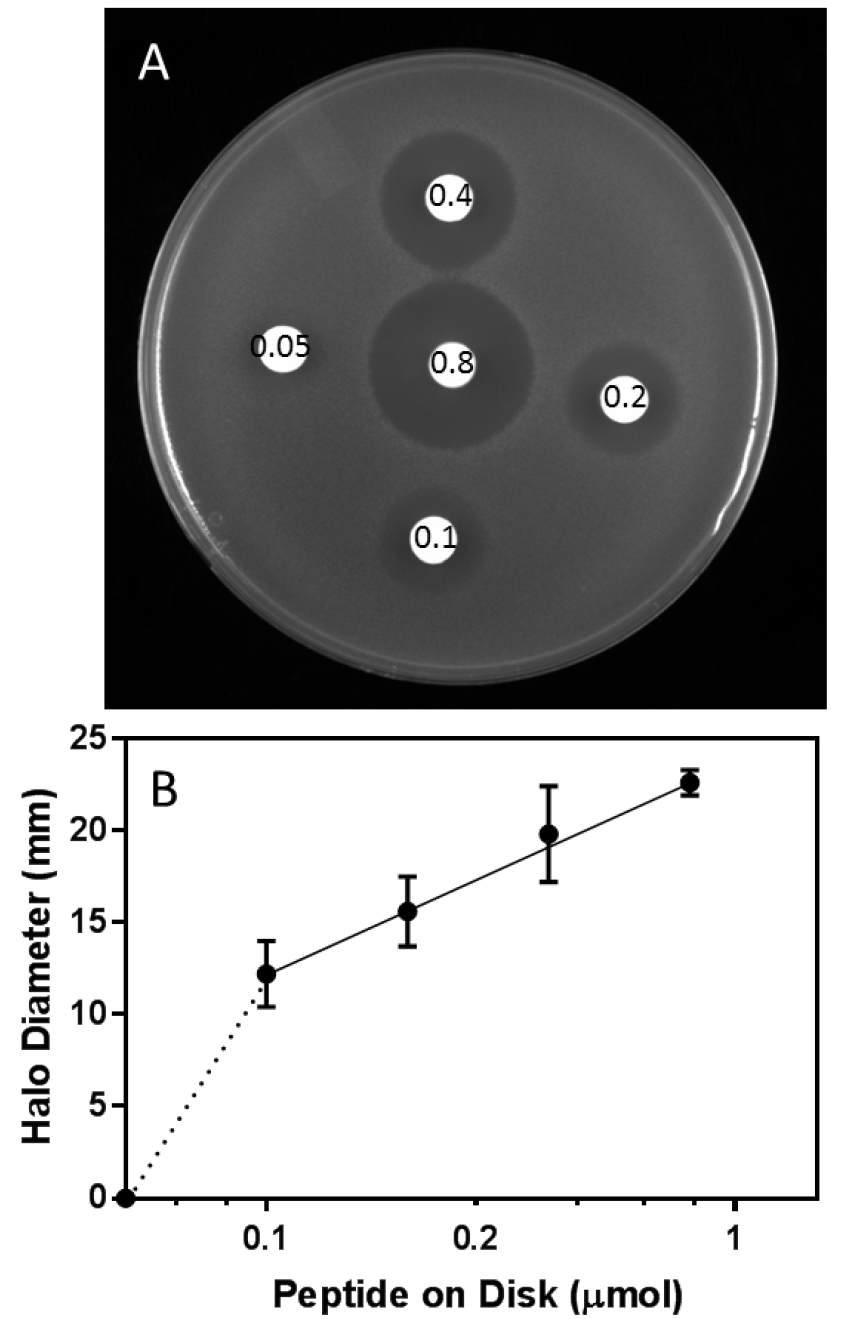
Figure 1. Halo assay and its analysis. A. Disks impregnated with the toxic peptide Ala-Eth (0.8-0.05 µmoles, as indicated) were distributed on a top agar lawn as shown and the Petri plates were incubated for 24 h at 30 °C. B. The halo diameter (mm) on each of three replicate plates was measured, averaged, and plotted as a function of the Log of peptide spotted onto the disk (µmoles).
Notes
For the halo assay: Ensuring that the top agar is evenly spread onto the growth plate is an essential factor for the development of a uniform zone of inhibition in the toxic halo assay.
Recipes
- Minimal proline (MP) broth (1 L)
20 g dextrose
1.7 g yeast nitrogen base (YNB) without (NH4)2SO4 and amino acids
1 g of proline as a nitrogen source
The dextrose is dissolved in 900 ml water and autoclave-sterilized. Once cooled, 100 ml of a filter sterilized 10x YNB-proline stock (17 g YNB and 10 g proline in 100 ml water) is added.
Note: Proline is used as a nitrogen source to induce the synthesis of the yeast peptide transport system. - MP+ broth
100 ml of MP broth
1.0 ml of uracil (2 mg/ml in double distilled water, filter-sterilized)
1.0 ml adenine sulfate (2 mg/ml 0.1 N HCl, filter-sterilized)
0.2 ml of histidine (10 mg/ml, filter-sterilized)
0.3 ml of leucine (10 mg/ml, filter-sterilized)
0.2 ml of tryptophan (10 mg/ml 0.1 N HCl, filter-sterilized)
Note: The above amino acids and nucleobases are added to satisfy the auxotrophic requirement of S. cerevisiae W303.The specific growth conditions may be modified for the yeast strain or toxic compound to be assayed. - MP+ growth plate
Dextrose (20 g) is dissolved in 900 ml water, 20 g of agar is added and this suspension is autoclave-sterilized
After the autoclaved solution is equilibrated to 55 °C, add the following solutions:
100 ml of filter-sterilized 10x YNB-proline
1.0 ml of uracil (2 mg/ml in double distilled water, filter sterilized)
1.0 ml adenine sulfate (2 mg/ml 0.1 N HCl, filter-sterilized)
0.2 ml of histidine (10 mg/ml, filter-sterilized)
0.3 ml of leucine (10 mg/ml, filter-sterilized)
0.2 ml of tryptophan (10 mg/ml 0.1 N HCl, filter-sterilized)
25 ml of this medium is aseptically poured into a 10 cm Petri plate
After solidification, the plates can be stored at room temperature for a week, or several weeks at 4 °C
Note: The above amino acids and nucleobases are added to satisfy the auxotrophic requirement of S. cerevisiae W303. - 1.1% Noble agar blanks
A solution of 1.1% Noble agar in water is prepared by bringing the solution up to boiling until all of the agar has dissolved.
The molten solution is dispensed into 16 x 125 mm glass screw cap tubes (3 ml per tube) and sterilized by autoclaving with the lids only partially screwed on.
Once cooled, the lids are tightened and the sealed tube stored for at least 6 months at 4 °C until needed.
Before use, tubes are immersed in boiling water to melt the agar then kept in a 55 °C water bath to cool, but maintain the liquid state prior to adding cells.
Acknowledgments
This protocol was adapted from our previous studies (Cai et al., 2006; 2007). This work was supported grants from the National Institute of General Medical Sciences.
References
- Cai, H., Hauser, M., Naider, F. and Becker, J. M. (2007). Differential regulation and substrate preferences in two peptide transporters of Saccharomyces cerevisiae. Eukaryot Cell 6(10): 1805-1813.
- Cai, H., Kauffman, S., Naider, F. and Becker, J. M. (2006). Genomewide screen reveals a wide regulatory network for di/tripeptide utilization in Saccharomyces cerevisiae. Genetics 172(3): 1459-1476.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Cai, H., Hauser, M., Naider, F. and Becker, J. M. (2016). Halo Assay for Toxic Peptides and Other Compounds in Microorganisms. Bio-protocol 6(22): e2025. DOI: 10.21769/BioProtoc.2025.
Category
Microbiology > Microbial cell biology > Cell-based analysis
Cell Biology > Cell-based analysis > Transport
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link