- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Acid Extraction of Total Histone from Colon Cancer HCT116 Cells
Published: Vol 6, Iss 22, Nov 20, 2016 DOI: 10.21769/BioProtoc.2023 Views: 11271
Reviewed by: HongLok LungXiaoyi ZhengJustine Marsolier

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Measuring Protein Synthesis during Cell Cycle by Azidohomoalanine (AHA) Labeling and Flow Cytometric Analysis
Koshi Imami and Tomoharu Yasuda
Apr 20, 2019 9752 Views

Analysis of the Ubiquitination and Phosphorylation of Vangl Proteins
Di Feng [...] Bo Gao
Oct 20, 2022 3421 Views

Isoform-specific, Semi-quantitative Determination of Highly Homologous Protein Levels via CRISPR-Cas9-mediated HiBiT Tagging
Kristina Seiler [...] Mario P. Tschan
Jul 20, 2023 2521 Views
Abstract
Histone acid extraction assay is a popular method to determine histone modification levels in mammalian cells. It includes three steps: first, histones are released from chromatin by sulfuric acid; trichloroacetate (TCA) is then added to precipitate histones; and finally, histones are dissolved in double-distilled H2O (ddH2O). Here we present a detailed histone acid extraction assay in our laboratory using a colon cancer cell line, HCT116, as a model.
Background
The nucleosome is the fundamental unit of eukaryotic chromatin, which is composed of a histone octamer (2 copies of H3, H4, H2A, H2B, respectively) wrapping by DNA (Strahl and Allis, 2000). The amino terminal of histone is subjected to a variety of post-translational modifications, such as methylation, acetylation, phosphorylation, ubiquitylation and sumoylation (Kouzarides, 2007). Although the function of these modifications has remained elusive, there is ever-growing studies suggest that histone modifications play vital roles in intracellular processes (Bannister and Kouzarides, 2011). Therefore, it is important to extract histones efficiently to detect histone modifications.
Histones can be extracted via different methods, in which histone acid extraction assay is one of the most popular procedures. It does not interrupt post-translational modifications of histones, and so it is very good for histone modification analysis. It has been tested that the extracted histones can be used in Western blot, and maybe other assays (not fully tested). However, immunoprecipitation is not recommended. In this protocol, we will present a detailed histone acid extraction assay, and describe how to release histones from chromatin, how to precipitate histones, and how to wash and dissolve histones in ddH2O.
Materials and Reagents
- 6 cm plate
- 1.5 ml tubes (Corning, Axygen®, catalog number: MCT-150-C )
- Human colon cancer cell line HCT116 (ATCC)
- Phosphate-buffered saline (PBS) (Thermo Fisher Scientific, GibcoTM)
- Acetone
- Double-distilled H2O (ddH2O)
- 2x SDS loading buffer (containing 200 mmol/L DTT)
- Tris-HCl (pH = 8.0)
- Potassium chloride (KCl)
- Magnesium chloride (MgCl2)
- Dithiothreitol (DTT)
- Protease inhibitor cocktail (Sigma-Aldrich, catalog number: P8340 )
- Sulfuric acid
- Trichloroacetate (TCA) (Sigma-Aldrich, catalog number: T6399 )
- Trichloroacetic acid (TCA) solution (see Recipes)
- Lysis buffer(see Recipes)
Equipment
- Pipettor (Eppendorf)
- Centrifuge (cooled and room-temperature)(Eppendorf)
- Rotator
- Spectrophotometer (Bio-Rad Laboratories)
- Metal bath
Procedure
An overview of the whole procedure is schematized in Figure 1.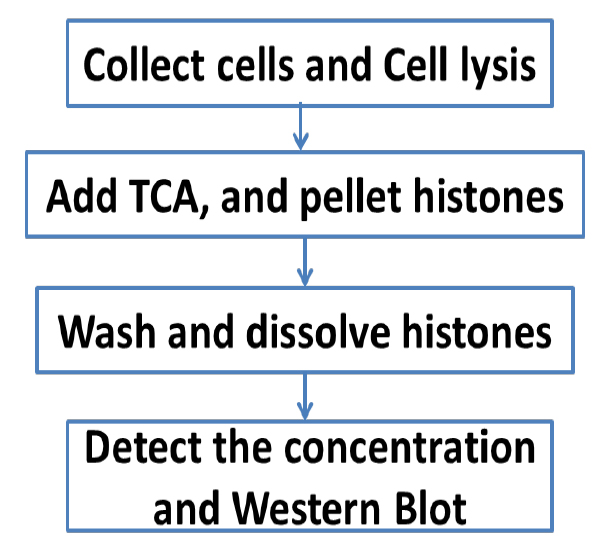
Figure 1. Scheme of the complete procedure described in the protocol
- Collect HCT116 cells (6 cm plate, 70-90% confluent) using any commonly used method in 1 ml ice-cold PBS and pellet (1,000 x g, 5 min, 4 °C).
- Discard the supernatant, add 1 ml ice-cold PBS and pipet gently to resuspend the cells, then pellet it again (1,000 x g, 5 min, 4 °C).
- Discard the supernatant, resuspend cell pellet in 400 μl lysis buffer, and incubate for 30 min with rotating at 4 °C.
- Pellet the intact nuclei by spinning in cooled centrifuge (12,000 x g, 10 min, 4 °C).
- Transfer the supernatant containing histones into a fresh 1.5 ml tube.
- Add 133 μl TCA drop by drop to histone solution and invert the tube several times to mix the solutions (final concentration of TCA is 25%). (It is better to add one drop and invert the tube at once to mix the solutions, and then add another drop.) The solution will appear milky over time.
- Incubate on ice for 30 min or overnight.
- Pellet histones by spinning in cooled centrifuge (12,000 x g, 10min, 4 °C).
- Carefully remove supernatant and wash histone pellet with 1 ml ice cold acetone without disturbing it. Acetone is used to remove acid from the solution without dissolving the histone pellet.
- Spin down in centrifuge at 12,000 x g, 10 min at 4 °C.
- Repeat steps 9 and 10 to wash histone pellet again.
- Carefully remove all of the supernatant and air-dry histone pellet for 30 min at room temperature.
- Dissolve histone pellet in appropriate volume of ddH2O (for example, 100 μl is appropriate for a 6 cm plate), and transfer into fresh 1.5 ml tube.
- Detect the concentration of histone solution with the spectrophotometer (using OD 280), and we may acquire 1-2 µg histone protein from a 70-90% confluent well on a 6 cm plate.
- Add the same volume of 2x SDS loading buffer into histone solution, and boil it on the metal bath (100 °C, 5 min), then subject to Western blot.
Representative data
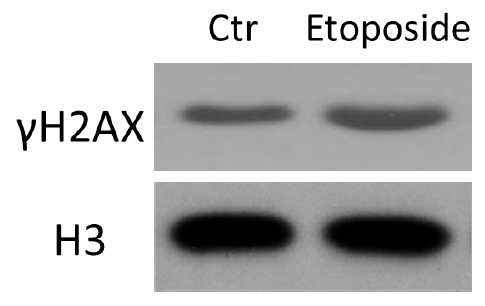
Figure 2. HCT116 cells were treated with etoposide and subjected to histone acid extraction. Immunoblotting was performed with the indicated antibodies.
Note: Relative Western blots have also been published in our paper Oncogene (Cao et al., 2016). For details, please refer to Figure 4 in Cao et al., 2016 (https://www.researchgate.net/publication/274316705_ATM-mediated_KDM2A_phosphorylation_is_required_for_the_DNA_damage_repair).
Recipes
- Trichloroacetic acid (TCA) solution
500 g TCA in 227 ml ddH2O
Store at room temperature, and protect from light - Lysis buffer
10 mM Tris-HCl, pH 8.0
1 mM KCl
1.5 mM MgCl2
10 mM DTT (added immediately before use)
1x protease inhibitor cocktail (added immediately before use)
0.4 M sulfuric acid (added immediately before use)
Acknowledgments
This work was supported by Beijing Natural Science Foundation grant 7164305, the ‘973 Projects’ (2011CB910100, 2011CB504200 and 2013CB911000), National Natural Science Foundation of China (81222028, 81321003, 81472581, 81530074, 31570812 and 91319302), and grants (B70001) from the Ministry of Science and Technology of China. This protocol was adapted from previous work published in Cancer Research (Zhu et al., 2001), Oncogene (Cao et al., 2016) and Oncotarget (Lu et al., 2015).
References
- Bannister, A. J. and Kouzarides, T. (2011). Regulation of chromatin by histone modifications. Cell Res 21(3): 381-395.
- Cao, L. L., Wei, F., Du, Y., Song, B., Wang, D., Shen, C., Lu, X., Cao, Z., Yang, Q., Gao, Y., Wang, L., Zhao, Y., Wang, H., Yang, Y. and Zhu, W. G. (2016). ATM-mediated KDM2A phosphorylation is required for the DNA damage repair. Oncogene 35: 301-313.
- Kouzarides T. (2007). Chromatin modifications and their function. Cell 128: 693-705.
- Lu, S., Yang, Y., Du, Y., Cao, L. L., Li, M., Shen, C., Hou, T., Zhao, Y., Wang, H., Deng, D., Wang, L., He, Q. and Zhu, W. G. (2015). The transcription factor c-Fos coordinates with histone lysine-specific demethylase 2A to activate the expression of cyclooxygenase-2. Oncotarget 6(33): 34704-34717.
- Strahl, B. D. and Allis, C. D. (2000). The language of covalent histone modifications. Nature 403(6765): 41-45.
- Zhu, W. G., Lakshmanan, R. R., Beal, M. D. and Otterson, G. A. (2001). DNA methyltransferase inhibition enhances apoptosis induced by histone deacetylase inhibitors. Cancer Res 61(4): 1327-1333.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Cao, L. and Zhu, W. (2016). Acid Extraction of Total Histone from Colon Cancer HCT116 Cells. Bio-protocol 6(22): e2023. DOI: 10.21769/BioProtoc.2023.
Category
Cancer Biology > General technique > Biochemical assays > Protein analysis
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









