- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Determining the Influence of Small Molecules on Hypoxic Prostate Cancer Cell (DU-145) Viability Using Automated Cell Counting and a Cell Harvesting Protocol
Published: Vol 6, Iss 22, Nov 20, 2016 DOI: 10.21769/BioProtoc.2017 Views: 8045
Reviewed by: Antoine de MorreeXiaoyi ZhengPooja Mehta

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Quantification of Autophagosomes in Human Fibroblasts Using Cyto-ID® Staining and Cytation Imaging
Barbara Hochecker [...] Jörg Bergemann
Jul 5, 2024 1660 Views

Microfluidic Cultures of Basal Forebrain Cholinergic Neurons for Assessing Retrograde Cell Death by Live Imaging
Srestha Dasgupta [...] Wilma J. Friedman
Jan 5, 2025 1860 Views

Real-time IncuCyte® Assay for the Dynamic Assessment of Live and Dead Cells in 2D Cultures
Arlene K. Gidda [...] Sharon M. Gorski
Feb 5, 2025 2865 Views
Abstract
Cell viability assays are an essential aspect of most cancer studies, however they usually require a considerable labor and time input. Here, instead of using the conventional microscopy and hemocytometer cell counting approach, we developed a cell harvesting protocol and combined it with the automated Countess Automated Cell Counter to generate cell viability data. We investigated the effects of dihydroxylated bile acids on the cell viability of prostate cancer cells grown under hypoxic conditions. We observed that for all conditions, cell viability was relatively unchanged, suggesting these molecules had little or no impact on cell viability. The combination of the automated approach and the cell harvesting protocol means this assay is i) easy to perform, ii) extremely reproducible and iii) it complements more conventional cancer assay data such as invasion, migration and adhesion.
Background
Determining the therapeutic utility of any biological molecule is a critical step in the development of novel molecular therapeutics to combat cancer progression and development. As a preliminary step in in vitro characterization, molecules must be assessed for their suitability as anti-cancer therapeutics. As part of this assessment, cell viability is a critical determinant of the cellular response to small molecules as it reflects the ability of a molecule to sustain a threshold cell viability, whilst simultaneously targeting key cancer progression mechanisms e.g., clonogenicity, invasion and adhesion. Conventional viability assays require an intensive labor input comprising microscopy, hemocytometers and manual cell counters. Here we developed a protocol for the rapid and accurate generation of cell viability data that will complement cancer research studies (Phelan et al., 2016).
Materials and Reagents
- 15 ml tubes (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 339650 )
- Blue 1 ml pipet tips (Greiner Bio One, catalog number: 686295 )
- Yellow 200 µl pipet tips (Greiner Bio One, catalog number: 739290 )
- Sterile Eppendorf tubes (Eppendorf, catalog number: 0030119401 )
- Disposable chamber slide
- DU-145 prostate cancer cells (ATCC, catalog number: HTB-81 )
- Dulbecco’s modified Eagle’s medium (DMEM) low glucose media (Sigma-Aldrich, catalog number: D5921-500ML )
- Deoxycholic acid (Sigma-Aldrich, catalog number: D2510-10G )
- 250 mM dimethyloxalylglycine (DMOG) in sterile PBS (EMD Millipore, Calbiochem, catalog number: 400091-50MG )
- Chenodeoxycholic acid (Sigma-Aldrich, catalog number: C9377-5G )
- Phosphate buffered saline (PBS) tablets (Sigma-Aldrich, catalog number: P4417-100TAB )
- Trypan blue (Thermo Fisher Scientific, GibcoTM, catalog number: 15250061 )
- Fetal bovine serum (FBS) (Sigma-Aldrich, catalog number: F6178-500ML )
- Penicillin-streptomycin (Sigma-Aldrich, catalog number: P4333-20ML )
- Glutamine (Sigma-Aldrich, catalog number: G7513-20ML )
- PBS solution (see Recipes)
- Complete Dulbecco’s media (see Recipes)
- 200 µM DMOG (see Recipes)
- CDCA and DCA bile acids (see Recipes)
Equipment
- T25 flasks (SARSTEDT, catalog number: 83.3910 )
- Cell scrapers (SARSTEDT, catalog number: 83.1831 )
- DM0412 centrifuge to accommodate 15 ml tubes (Scilogex, model: DM0412 )
- Water jacketed CO2 incubator (Thermo Fisher Scientific, Thermo ScientificTM, model: Series II )
- Automated pipette filler (Gilson, catalog number: F110753 )
- Countess automated cell counter (Thermo Fisher Scientific, InvitrogenTM, catalog number: C10227 )
- Inverted microscope (OLYMPUS, model: CKX31 )
- Disposable cell chamber slides (Thermo Fisher Scientific, InvitrogenTM, catalog number: C10228 )
- Hemocytometer (Thermo Fisher Scientific, Fisher Scientific, catalog number: 10350141 )
Software
- Excel
Procedure
- Grow DU-145 prostate cancer cells under hypoxic conditions using 200 μM DMOG; DMOG upregulates normoxic HIF-1α by inhibiting HIF-1 prolyl and asparaginyl hydroxylase function at the oxygen dependent degradation domain (ODDD) of the protein. Seed cells at a density of 50,000 cells into T25 flasks in complete Dulbecco’s media and allow them to become confluent over 1-2 days prior to experimentation. Once confluent, remove the media and treat cells with media containing 100 μM bile acids + 200 μM DMOG, DMOG only and media only (untreated controls). Generate enough flasks to ensure viability assessment every 24 h over a 5 day period.
- After 24 h, on day 1 of the assay, decant the media, wash the flask surface in sterile PBS to remove debris and gently scrape cells from the flask surface using a cell scraper. Avoid trypsin-EDTA due to potential cytotoxic effects. Add 2 ml of sterile PBS to the flask to create a cell suspension. Mix thoroughly (by gently pipetting up and down) to homogenize cell clumps.
- Transfer this 2 ml volume to a 15 ml Falcon tube and centrifuge at 160 x g for 2 min to pellet cells.
- Do not fully decant the supernatant, leave approximately 100 µl at the bottom of the tube. Mix this volume gently with the pellet, using a 1 ml blue pipet tip to yield an approximate cell suspension of 100-200 µl.
- Add 20 μl of the cell suspension and 20 μl of trypan blue into a new Eppendorf tube and pipette mix to completely homogenize clumps. Flick mix for a few seconds in order to stain cells.
- Load two 10 μl aliquots onto each side of a disposable chamber slide (reusable) and place into the CountessTM automated cell counter to enumerate live/dead cells (viability), represented as percentage cell viability.
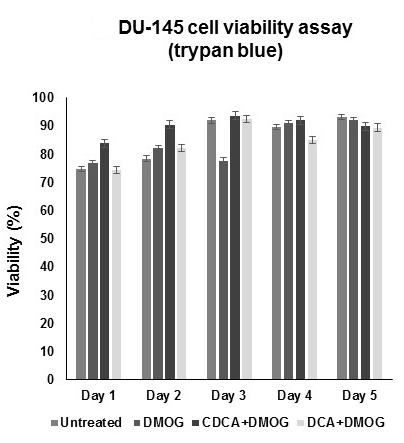
Figure 1. Dihydroxylated bile acids CDCA and DCA do not affect the cell viability of hypoxic DU-145 cells. For cell viability (trypan blue) assays, all cells were grown under hypoxic conditions (DMOG 200 μM), except for untreated cells. No significant compromises were observed in cell viability with respect to time. All data were derived from three independent experiments with at least three replicates in each experiment. Standard deviation was used to generate error bars. - Repeat the procedure each day until data for all 5 days have been collected.
Percentage viability data can be read directly from the automated cell counter. Graph data as viability (%) versus time (day) using Excel (Figure1).
Notes
Percentage viability data can be read directly from the automated cell counter. Graph data as viability (%) versus time (day) using Excel (Figure1).
Data analysis
- Trypsin-EDTA should not be used in this protocol as it may induce some cell death which may affect viability thus distorting the data.
- Only the DU-145 prostate cancer cell line was tested using this method.
- Disposable cell chamber slides can be used 4-5 times (with washing in water) however with time, they become less efficient (bubbles develop in the counting area, distorting counting).
- While this assay was carried out in DU-145 prostate cancer cells grown under hypoxic conditions, this assay could be adapted, with some trouble-shooting to those cancer cells grown under normoxic conditions. Indeed, for this assay, untreated DU-145 cells were grown under normoxic conditions and yielded similar viabilities to those of hypoxic test cells (DMOG alone), suggesting hypoxia had little or no impact on viability. The assay could be applicable to any adherent cell line, grown under normoxia or hypoxia conditions, where cell viability is the main measurement output.
- To corroborate cell counter data, a hemocytometer was used to manually count cells from random flasks over the duration of the assay (5 days). Overall, an average cell viability of 93.56% ± 2% was recorded suggesting relative parity between methods.
- As long as cells are adherent, they can be grown on any sized plate/dish/flask. However, technical issues involving cell scraping may arise on low surface areas.
- DU-145 prostate cancer cells are adherent spindle shaped epithelial cells that require passaging every 2-3 days. Typically, cells can be routinely passaged up to passage 20.
Recipes
- PBS solution
Dissolve 5 PBS tablets in 1 L distilled water and autoclave - Complete Dulbecco’s media
10% fetal bovine serum
0.5% penicillin-streptomycin
2 mM glutamine - 200 µM DMOG
Add 1.15 ml sterile PBS to desiccated powder in bottle
Mix thoroughly and use at a final concentration of 200 µM - CDCA and DCA bile acids
50 mM stock solution in distilled water
Final concentration used: 100 µM
Acknowledgments
This research was supported in part by grants awarded by the European Commission, Science Foundation Ireland, the Department of Agriculture and Food, Ireland, the Irish Research Council for Science, Engineering and Technology and the Health Research Board.
References
- Phelan, J. P., Reen, F. J., Dunphy, N., O'Connor, R. and O'Gara, F. (2016). Bile acids destabilise HIF-1α and promote anti-tumour phenotypes in cancer cell models. BMC Cancer 16: 476.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Phelan, J. P., Reen, F. J. and O’Gara, F. (2016). Determining the Influence of Small Molecules on Hypoxic Prostate Cancer Cell (DU-145) Viability Using Automated Cell Counting and a Cell Harvesting Protocol. Bio-protocol 6(22): e2017. DOI: 10.21769/BioProtoc.2017.
Category
Cancer Biology > Cell death > Biochemical assays
Cell Biology > Cell viability > Cell death
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










