- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Analysis of Phagosomal Antigen Degradation by Flow Organellocytometry
Published: Vol 6, Iss 22, Nov 20, 2016 DOI: 10.21769/BioProtoc.2014 Views: 9242

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation and Ex Vivo Testing of CD8+ T-Cell Division and Activation Using Mouse Splenocytes
Melissa Dolan [...] John M.L. Ebos
Aug 20, 2025 3878 Views

Detection of Autophagy in Human Peripheral Blood Mononuclear Cells Using Guava® Autophagy and Flow Cytometry
Melanie Scherer [...] Jörg Bergemann
Sep 20, 2025 1418 Views

Protocol for the Isolation and Analysis of Extracellular Vesicles From Peripheral Blood: Red Cell, Endothelial, and Platelet-Derived Extracellular Vesicles
Bhawani Yasassri Alvitigala [...] Lallindra Viranjan Gooneratne
Nov 5, 2025 1448 Views
Abstract
Professional phagocytes internalize self and non-self particles by phagocytosis to initiate innate immune responses. After internalization, the formed phagosome matures through fusion and fission events with endosomes and lysosomes to obtain a more acidic, oxidative and hydrolytic environment for the degradation of its cargo. Interestingly, phagosome maturation kinetics differ between cell types and cell activation states. This protocol allows to quantify phagosome maturation kinetics on a single organelle level in different types of phagocytes using flow cytometry. Here, ovalbumin (OVA)-coupled particles are used as phagocytosis model system in dendritic cells (DC), which are internalized by phagocytosis. After different time points, phagosome maturation parameters, such as phagosomal degradation of OVA and acquisition of lysosomal proteins (like LAMP-1), can be measured simultaneously in a highly quantitative manner by flow organellocytometry. These read-outs can be correlated to other phagosomal functions, for example antigen degradation, processing and loading in DC.
Background
In innate immunity, professional phagocytes such as dendritic cells (DC), macrophages and neutrophils recognize and internalize different types of particles by phagocytosis including pathogens and dead cells (Flannagan et al., 2012). Intra-phagosomal degradation of these particles by fission and fusion events with endosomal and lysosomal compartments allow either clearance and complete destruction of phagosomal cargo or partial degradation and processing of phagosomal antigens for presentation to T lymphocytes. Different parameters of phagosome maturation, such as acidification, oxidation and proteolysis, dictate phagosomal fate and influence the initiation of different immune responses (Kinchen and Ravichandran, 2008). In particular, the type of involved phagocytes, specific recognition of pathogen-associated molecular patterns (such as LPS) or danger-associated molecular patterns (such as HMGB1) on the surface of the particle as well as the influence of cellular and phagosomal signal transduction determine strength and duration of phagosomal antigen degradation (Savina and Amigorena, 2007).
This protocol was developed to follow antigen degradation kinetics on the single phagosome level in a highly quantitative fashion by flow organellocytometry. It is based on a method previously published by our lab using antigen-coupled polystyrene beads as phagocytosis model system (Savina et al., 2010). Due to their physical properties, bead-containing phagosomes can be distinguished from other cell organelles of similar size during flow cytometry. Therefore, phagosomal antigen degradation can be measured directly without previous organelle fractionation and purification methods. Another major advantage of this protocol over many other protocols is the fact that it allows to distinguish between internalized beads within phagosomes and particles bound on the outside of the cell, which were not phagocytosed. This protocol was used previously for the characterization of phagosomal antigen degradation in bone marrow-derived DC (BMDC) (Hoffmann et al., 2012; Alloatti et al., 2015) as well as in splenic DC (Alloatti et al., 2015). However, other phagocyte types and different antigen sources can be used as well for the investigation of antigen degradation kinetics in phagosomes. The approach described below is adapted to BMDC and ovalbumin (OVA)-coupled particles as phagosomal cargo.
Materials and Reagents
Note: The entire method is performed with sterile pyrogen-free dishes and plates, pipettes, tips, microfuge and conical tubes. All media and buffers need to be filtered through 0.22 μm filters.
- Petri dish, 145 x 20 mm (Greiner Bio One, catalog number: 639161 )
- 2 ml tubes
- 15 ml centrifuge tube
- U-bottom 96-well storage plate (Corning, Falcon®, catalog number: 353077 )
- V-bottom 96-well storage plate (Corning, Falcon®, catalog number: 353263 )
- 2 ml sterile syringe (Henke Sass Wolf, catalog number: 4020.000V0 )
- 22 G sterile needle, 0.7 x 40 mm (Terumo Europe, catalog number: NN-2238R )
- Mice: C57BL/6J (Janvier Labs)
- Particles: amine-modified polystyrene microspheres, 3 μm diameter (Polysciences, catalog number: 17145-5 )
- Dulbecco’s phosphate-buffered saline, no calcium, no magnesium (DPBS) (Thermo Fisher Scientific, GibicoTM, catalog number: 14190094 )
- Glutaraldehyde, 25% (vol/vol), EM grade (Electron Microscopy Sciences, catalog number: 16220 )
- Low endotoxin ovalbumin (OVA) (Worthington Biochemical, catalog number: LS003062 )
- Glycine, 0.5 M in PBS (Biosolve, catalog number: 07132391 )
- CO2-independent medium (Thermo Fisher Scientific, GibcoTM, catalog number: 18045088 )
- Glutamax supplement (100x) (Thermo Fisher Scientific, GibcoTM, catalog number: 35050061 )
- Iscove’s modified Dulbecco’s medium (IMDM) (Thermo Fisher Scientific, GibcoTM, catalog number: 31980030 )
- Low endotoxin fetal bovine serum (FBS, heat-inactivated for 20 min at 56 °C) (Biowest, catalog number: S1860 )
- β-mercaptoethanol (50 mM) (Thermo Fisher Scientific, GibcoTM, catalog number: 31350010 )
- Penicillin-streptomycin (10,000 U/ml) (Thermo Fisher Scientific, GibcoTM, catalog number: 15140122 )
- Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A4503 )
- Anti-chicken egg albumin (OVA antibody) (Sigma-Aldrich, catalog number: C6534 )
- Purified rat anti-mouse CD16/CD32 monoclonal antibody (Fc block) (BD, PharmingenTM, catalog number: 553142 )
- Anti-mouse LAMP-1 antibody, biotin conjugate (Affymetrix, eBioscience, catalog number: 13-1071-82 )
- Goat anti-rabbit IgG (H+L) antibody, DyLight 633 conjugate (Thermo Fisher Scientific, InvitrogenTM, catalog number: 35562 )
- Goat anti-rabbit IgG (H+L) antibody, Alexa Fluor 568 conjugate (Thermo Fisher Scientific, InvitrogenTM, catalog number: A11036 )
- Streptavidin, Alexa Fluor® 488 conjugate (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: S11223 )
- Imidazole (Sigma-Aldrich, catalog number: I-0250 )
- Sucrose (EMD Millipore, catalog number: 107651 )
- Phenylmethanesulfonyl fluoride (PMSF) (Sigma-Aldrich, catalog number: P-7626 )
- EDTA-free protease inhibitor cocktail (Sigma-Aldrich, catalog number: 11873580001 )
- Dithiothreitol (DTT) (EMD Millipore, catalog number: 233155 )
- Trypan blue solution, 0.4% (wt/vol) (MP Biomedicals, catalog number: 0916910 )
- Internalization medium (see Recipes)
- BMDC culture medium (see Recipes)
- Homogenization buffer (see Recipes)
Equipment
- Vortex
- Refrigerated centrifuge for tubes of 2 ml, 15 ml and 50 ml size as well as 96-well plates
- Tissue culture incubator adjusted to 37 °C and 5% CO2
- Stuart test tube rotator wheel tolerating 4 °C (Bibby Scientific, model: SB3 )
- Temperature-controlled water bath
- Pipets
- Tissue culture light microscope equipped with bright field and 20x objective
- Multi-channel pipet
- LSR II flow cytometer (BD) or any other multicolour flow cytometer
Software
- FlowJo software (FlowJo, LLC.)
- Prism 6 software (GraphPad Software, Inc.)
Procedure
- Preparation of antigen-coupled particles
Note: Estimate the number of cells, which will be used for this assay, in advance to prepare a sufficient amount of particles coupled to OVA the day before the experiment. The stock suspension of 3 μm microspheres contains 1.68 x 109 particles/ml.
Example: Estimate the amount of cells you will harvest based on the amount of dishes in culture (for example 20 x 106 cells in total from four 145 mm dishes of BMDC). Multiply this number by 10 (the amount of beads provided per cell) and divide by the concentration of beads (1.68 x 109 particles/ml).
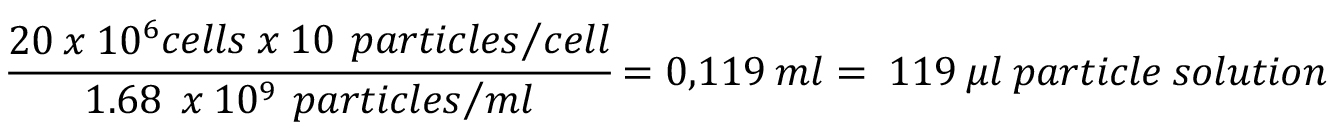
- Vortex suspension of amine-modified polystyrene microspheres thoroughly and distribute into 2 ml tubes (max. 200 μl/tube).
- Add 1.6 ml PBS to each tube, vortex and spin for 3 min at 16,000 x g at 4 °C.
- Discard supernatant, resuspend the pellet in 1.8 ml PBS and vortex. Spin again.
- Discard supernatant and resuspend the pellet in 8% glutaraldehyde/PBS (vol/vol). Use five times the volume of beads, which was used initially (e.g., 200 μl in 1 ml).
- Vortex and combine all suspensions in a 15 ml centrifuge tube. Protect the tube from light and incubate for 4 h on a test-tube rotator at 20 rpm at room temperature.
- Transfer suspension back into 2 ml tubes and spin them for 3 min at 16,000 x g at 4 °C.
- Discard supernatant, resuspend and vortex pellet in 1.8 ml PBS. Spin again.
- Discard supernatant and resuspend the pellet in 0.5 mg/ml OVA/PBS (Use five times the volume of beads that was used initially [e.g., 200 μl in 1 ml]).
- Vortex and combine all suspensions in a 15 ml centrifuge tube. Incubate overnight on a test-tube rotator at 20 rpm at 4 °C.
- Transfer suspension back into 2 ml tubes and spin them for 3 min at 16,000 x g at 4 °C.
- Discard supernatant, resuspend and vortex pellet in 1.8 ml 0.5 M glycine/PBS.
- Incubate for 30 min on a test-tube rotator at 20 rpm at 4 °C.
- Spin tubes for 3 min at 16,000 x g at 4 °C.
- Discard supernatant, resuspend and vortex pellet in 1.8 ml PBS.
- Repeat the spin followed by two more washes with 1.8 ml PBS (a total of three washes).
- Resuspend each pellet in the initial volume (200 μl), pool and vortex the dispersion. Store the suspension of OVA-coupled microspheres at 4 °C.
Note: Coupled microspheres should be prepared freshly the day before the flow organellocytometry experiment and can be used within a week when stored at 4 °C. Never freeze particles and always vortex dispersion carefully to avoid aggregation of coupled microspheres.
- Vortex suspension of amine-modified polystyrene microspheres thoroughly and distribute into 2 ml tubes (max. 200 μl/tube).
- Phagocytosis of antigen-coupled particles
Note: Estimate around 5 x 106 cells for each experimental condition.
- Pick up cell clusters and single cells by resuspending them with a pipet. Spin cells for 4 min at 400 x g at 4 °C.
- Resuspend cells in ice-cold PBS and spin them again for 4 min at 400 x g at 4 °C.
- Resuspend cells in ice-cold internalization medium, transfer them to a conical 15 ml centrifuge tube and count the cells.
- Adjust the cell suspension in ice-cold internalization medium to a cell density of 20 x 106/ml and keep them on ice.
- Vortex suspension of OVA-coupled microspheres and add them to the cells at a particle-to-cell ratio of 10:1. Mix carefully.
- Incubate samples for 30 min in a water bath adjusted to 16 °C. If necessary, add ice to the water bath during the incubation period.
- Place samples on ice and add 10 ml ice-cold PBS to the tubes. Pipet up and down, close the tubes and shake them thoroughly for 10 sec. Spin tubes for 4 min at 100 x g at 4 °C to remove floating particles.
- Discard supernatant and repeat the last step twice to have a total of three washes.
- Resuspend each cell pellet in pre-warmed BMDC culture medium and divide cell suspension in 15 ml centrifuge tubes to allow phagosomal antigen degradation to occur for different time points. Use 1-2 ml medium for each experimental condition.
- Add 10 ml ice-cold PBS to one tube, which represents the time point of 0 min. Keep this tube on ice.
- Keep the other tubes open and incubate them at 37 °C and 5% CO2 for different chase periods (for example: 60 and 120 min).
- Stop each chase period by adding 10 ml of ice-cold PBS and keep these tubes on ice.
- Pick up cell clusters and single cells by resuspending them with a pipet. Spin cells for 4 min at 400 x g at 4 °C.
- Manual lysis and labeling of samples
Note: After phagocytic uptake of particles, cells undergo a labeling step that distinguishes surface-bound particles from internalized ones. Subsequently, cells are lysed manually to release cytosol and cell organelles followed by a labeling step to detect levels of phagosomal OVA as well as the acquisition of lysosomal markers (for example, LAMP-1) to phagosomes during phagosome maturation.
- Spin all tubes for 4 min at 400 x g at 4 °C.
- Discard supernatant and resuspend each pellet in PBS + 1% (wt/vol) BSA. Transfer samples to a U-bottom 96-well plate.
- Spin the plate for 4 min at 400 x g at 4 °C and flick the supernatant.
- Resuspend each sample in 0.2 ml PBS + 1% (wt/vol) BSA + 1:100 diluted CD16/CD32 antibody (Fc block). Incubate on ice for 10 min.
- Spin the plate for 4 min at 400 x g at 4 °C and flick the supernatant.
- Resuspend each sample in 0.2 ml PBS + 1% (wt/vol) BSA + 1:500 diluted OVA antibody. Incubate on ice for 15 min.
- Spin the plate for 4 min at 400 x g at 4 °C and flick the supernatant.
- Resuspend each sample in 0.2 ml PBS and spin the plate for 4 min at 400 x g at 4 °C. Flick the supernatant and repeat the wash.
- Resuspend each sample in 0.2 ml PBS + 1% (wt/vol) BSA + 1:1,000 diluted anti-rabbit Alexa Fluor 568. Incubate on ice for 15 min.
- Spin the plate for 4 min at 400 x g at 4 °C and flick the supernatant.
- Resuspend each sample in 0.2 ml PBS and spin the plate for 4 min at 400 x g at 4 °C. Flick the supernatant and repeat the wash.
- Transfer samples into conical 1.5 ml centrifuge tubes and spin them for 4 min at 400 x g at 4 °C.
- Aspirate the supernatant carefully and resuspend each cell pellet in 0.5 ml homogenization buffer.
- For mechanical lysis of the cells, pass the suspension 15 times thoroughly through a 22 G needle fitted to a 2 ml syringe. Use the whole volume of the syringe for mechanical breaking of cells. Control homogenization success by trypan blue staining (Figure 1): Add 1 μl sample to 9 μl 0.4% (wt/vol) trypan blue solution, mix and add to a cell culture counting chamber. Count the amount of unstained, intact cells as well as stained, lysed cells using a light microscope. Calculate the percentage of lysed cells and do not exceed 80% of lysed cells. Additionally, avoid breaking of cell nuclei.
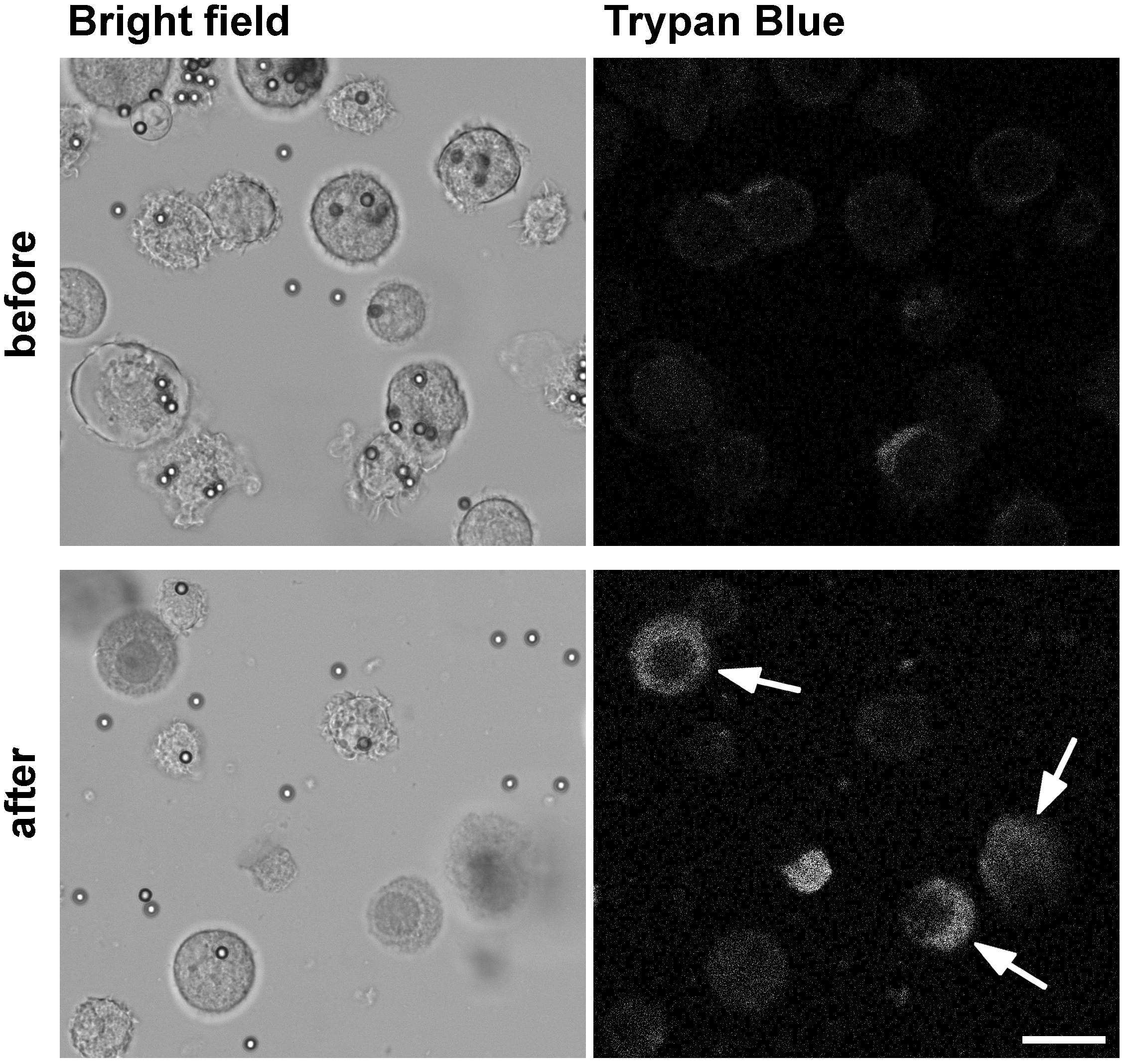
Figure 1. Mechanical lysis of BMDC after uptake of OVA-coupled beads to release phagosomes for subsequent flow organellocytometry. BMDC were allowed to internalize OVA-coupled beads for 30 min followed by a chase period to allow phagosome maturation to occur. To break cells mechanically in order to release intact phagosomes without breaking cell nuclei, we pass the cell suspension 15 times through a 22 G needle. The image shows the cell suspension labeled with trypan blue before (upper panel) and after cell lysis (lower panel). Accumulation of trypan blue in cells with broken cell membrane but intact nuclei (arrows) allows one to monitor success of mechanical cell lysis. The release of phagosomes in the obtained post-nuclear supernatant (PNS) is visualized by bright field microscopy. Scar bar = 20 μm.
- Spin the tubes for 4 min at 150 x g at 4 °C to separate the post-nuclear supernatant (PNS) from cell nuclei and remaining intact cells.
- Transfer the PNS of the different samples into a V-bottom 96-well plate and keep on ice.
- Spin the plate for 3 min at 1,500 x g at 4 °C and remove supernatant with a multi-channel pipet.
- Add 50 μl PBS + 1% (wt/vol) BSA + 1:500 diluted OVA antibody + 1:100 diluted LAMP-1 antibody.
- Mix samples with a multi-channel pipet, seal the plate and incubate overnight on ice.
- Add 0.15 ml PBS + 0.1% (wt/vol) BSA to each well, spin the plate for 3 min at 1,500 x g at 4 °C and remove supernatant with a multi-channel pipet.
- Add 0.2 ml PBS + 0.1% (wt/vol) BSA and mix samples with a multi-channel pipet.
- Spin the plate again and repeat the wash with PBS + 0.1% (wt/vol) BSA once more.
- After the supernatant is removed with a multi-channel pipet, add 50 μl PBS + 1% (wt/vol) BSA + 1:1,000 diluted anti-rabbit DyLight 633 + 1:1,000 diluted streptavidin Alexa Fluor 488. Incubate on ice for 45 min.
- Add 0.15 ml PBS + 0.1% (wt/vol) BSA to each well, spin the plate for 3 min at 1,500 x g at 4 °C and remove supernatant with a multi-channel pipet.
- Add 0.2 ml PBS + 0.1% (wt/vol) BSA and mix samples with a multi-channel pipet.
- Spin the plate again and repeat the wash with PBS + 0.1% (wt/vol) BSA once more.
Resuspend each sample in 0.2 ml PBS and keep the plate on ice until the measurement by flow cytometry. Measure the samples in a non-fixed state on the same day.
- Spin all tubes for 4 min at 400 x g at 4 °C.
Data analysis
Note: After the PNS, which contains the phagosomes, has been labeled for OVA and LAMP-1, it is analyzed by flow organellocytometry to determine the kinetics of antigen degradation (level of phagosomal OVA at a given time point) and of the acquisition of lysosomal markers (e.g., LAMP-1). Due to the physical properties and the specific size of microsphere-containing phagosomes, a gating strategy can be applied to measure these parameters simultaneously.
- Measure the samples by multicolour flow cytometry and apply the following gating strategy:
- Determine the population of beads and phagosomes in the PNS by measuring OVA-coupled beads alone and apply similar forward scatter (FSC) and side scatter (SSC) settings to your samples. Set a gate on the single particle population (Figure 2A).
- Exclude surface-bound beads (that are labeled before manual lysis) from subsequent analysis. Include only phagosomes, which are negative for OVA detected by anti-rabbit Alexa 568, by applying a second gate (Figure 2B).
- The gated population contains single phagosomes, which can be analyzed over time for OVA degradation and LAMP-1 acquisition either simultaneously (Figure 2C) or separately (Figure 2D) by plotting histograms of OVA (detected by anti-rabbit DyLight 633) and LAMP-1 (detected by streptavidin Alexa Fluor 488).
Note: Depending on the applied fluorophores for antibody labeling and the used flow cytometer, compensation between the different channels might be necessary.
- Analysis of flow cytometry data was performed using FlowJo software (FlowJo, LLC.). Statistical analysis was performed using Prism 6 software (GraphPad Software, Inc.). Each single experiment needs to be repeated by a sufficient number of independent experiments (at least in triplicate) to draw conclusions from the flow organellocytometry analysis. The data shown in Figure 2 is representative of at least three independent experiments.
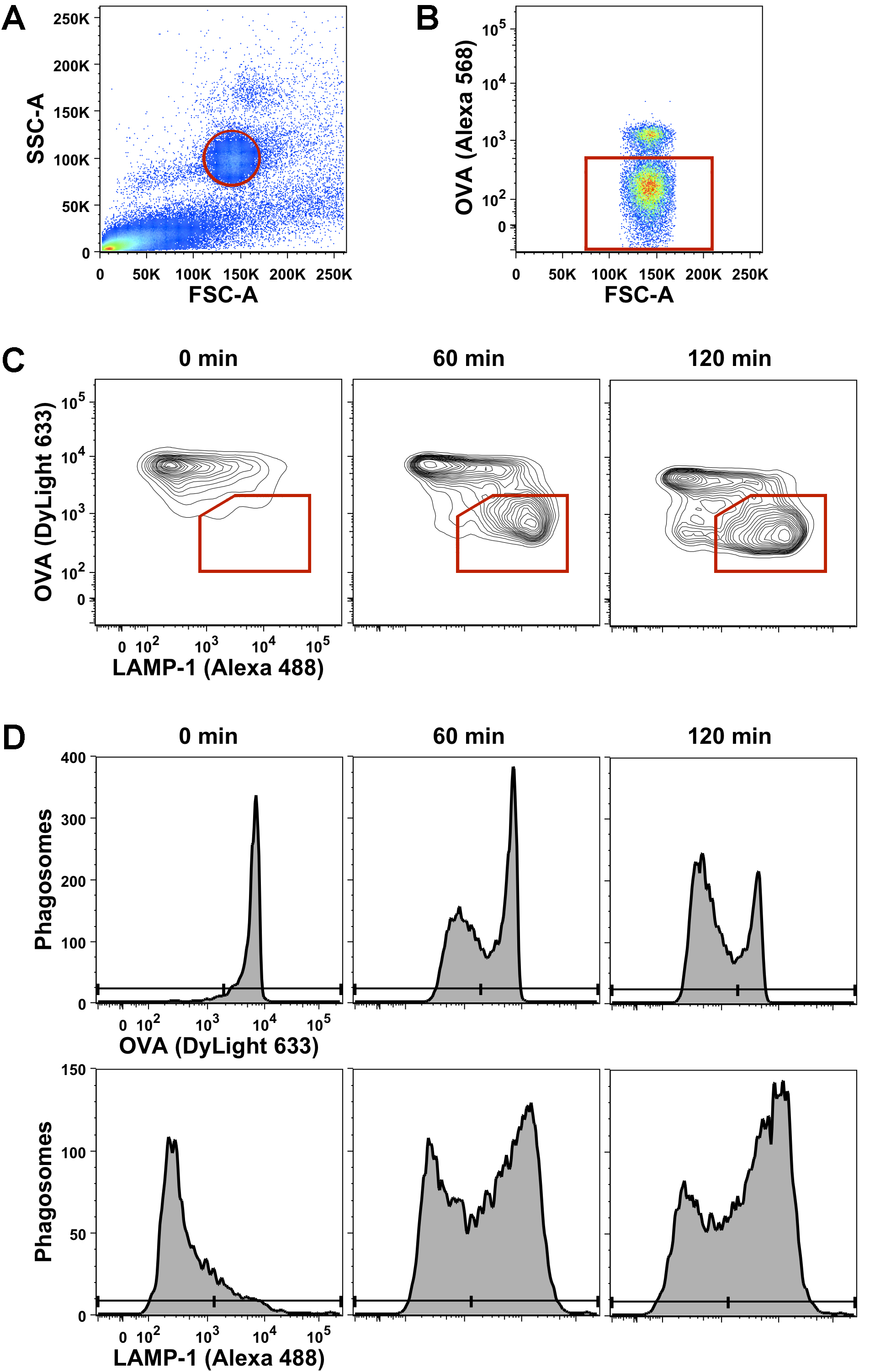
Figure 2. Gating strategy of the applied flow organellocytometry analysis of DC phagosomes. LPS-treated BMDC were allowed to internalize OVA-coated beads at 16 °C and incubated for different chase periods (0 min, 60 min, 120 min) at 37 °C to allow phagosome maturation. Surface-bound, non-internalized beads were stained with anti-OVA and an Alexa 568-coupled secondary antibody. Subsequently, cells were mechanically lysed to release the post-nuclear supernatant (PNS) including bead-containing phagosomes. The PNS was stained for OVA (detected by a DyLight 633-coupled secondary antibody) and LAMP-1 (detected by an Alexa 488-coupled secondary antibody) and analyzed simultaneously by flow cytometry. A. The first gate was set on single beads and single bead-containing phagosomes using forward scatter (FSC) and side scatter (SSC) settings of the labeled PNS. B. In a second gate, non-internalized beads (positive for Alexa 568) were separated from bead-containing phagosomes (negative for Alexa 568). C. Phagosomes were analyzed simultaneously for degradation of OVA and acquisition of LAMP-1 by applying a third gate on the mature, OVA (Dylight 633)-negative and LAMP-1 (Alexa 488)-positive population. D. Alternatively, these two parameters of phagosome maturation can also be analyzed separately and quantified over time.
Notes
- BMDC are generated from isolated murine bone marrow progenitor cells in GM-CSF-containing medium for 9 days. On the day of experiment, cell surface expression of CD11c should always exceed 85%.
- The reproducibility of results is highly dependent on homogenous conditions during mechanical breaking and cell lysis. Always control the success of cell lysis by trypan blue staining. Another factor is the phagocytic efficiency of the used cells. Always count cell numbers of your samples to apply comparable bead-to-cell ratios during bead uptake. Do not exceed this ratio, because high numbers of internalized particles will induce cell death and influence the parameters that are measured by flow organellocytometry.
- Although OVA-coupled particles are used here as phagocytosis model system, other ligands can be applied during bead coupling to investigate the influence of receptor-ligand interactions on phagosome maturation. Some examples are published elsewhere (Hoffmann et al., 2010; Hoffmann et al., 2012).
Recipes
- Internalization medium
CO2-independent medium
1x glutamax supplement
- BMDC culture medium
IMDM
10% (vol/vol) FBS
50 μM β-mercaptoethanol
100 IU/ml penicillin
100 μg/ml streptomycin
10% (vol/vol) supernatant from J558 plasmacytoma cells, which was used as GM-CSF source (Winzler et al., 1997)
- Homogenization buffer
3 mM imidazole, pH 7.4
250 mM sucrose
2 mM PMSF
1x protease inhibitor cocktail
2 mM DTT
Acknowledgments
This work was supported by the French National Research Agency through the ‘Investments for the Future’ program (France-BioImaging, ANR-10-INSB-04), ANR-11-LABX-0043 and by the CelTisPhyBio Labex (N- ANR-10-LBX-0038), part of the IDEX PSL (ANR-10-IDEX-0001-02 PSL). We are grateful to the financial support by the European Research Council (2013-AdG No.340046 DCBIOX), by La Ligue Nationale contre le Cancer (EL2014.LNCC/SA), by Fonds Wetenschappelijk Onderzoek (FWO; 1526615N; 11W8415N), by an EMBO long-term fellowship (ALTF 883-2011) and by fellowships of Fondation Recherche Médicale (SPF20101221176) and the omics@VIB program (co-financed by the Marie Curie FP7 People Cofund).
The protocol described here is based on a previously published protocol by our lab (Savina et al., 2010), which was developed further to measure different phagosome maturation parameters in dendritic cells and macrophages.
References
- Alloatti, A., Kotsias, F., Pauwels, A. M., Carpier, J. M., Jouve, M., Timmerman, E., Pace, L., Vargas, P., Maurin, M., Gehrmann, U., Joannas, L., Vivar, O. I., Lennon-Dumenil, A. M., Savina, A., Gevaert, K., Beyaert, R., Hoffmann, E. and Amigorena, S. (2015). Toll-like receptor 4 engagement on dendritic cells restrains phago-lysosome fusion and promotes cross-presentation of antigens. Immunity 43(6): 1087-1100.
- Flannagan, R. S., Jaumouille, V. and Grinstein, S. (2012). The cell biology of phagocytosis. Annu Rev Pathol 7: 61-98.
- Hoffmann, E., Kotsias, F., Visentin, G., Bruhns, P., Savina, A. and Amigorena, S. (2012). Autonomous phagosomal degradation and antigen presentation in dendritic cells. Proc Natl Acad Sci U S A 109(36): 14556-14561.
- Hoffmann, E., Marion, S., Mishra, B. B., John, M., Kratzke, R., Ahmad, S. F., Holzer, D., Anand, P. K., Weiss, D. G., Griffiths, G. and Kuznetsov, S. A. (2010). Initial receptor-ligand interactions modulate gene expression and phagosomal properties during both early and late stages of phagocytosis. Eur J Cell Biol 89(9): 693-704.
- Kinchen, J. M. and Ravichandran, K. S. (2008). Phagosome maturation: going through the acid test. Nat Rev Mol Cell Biol 9(10): 781-95.
- Savina, A. and Amigorena, S. (2007). Phagocytosis and antigen presentation in dendritic cells. Immunol Rev 219: 143-156.
- Savina, A., Vargas, P., Guermonprez, P., Lennon, A. M. and Amigorena, S. (2010). Measuring pH, ROS production, maturation, and degradation in dendritic cell phagosomes using cytofluorometry-based assays. Methods Mol Biol 595: 383-402.
- Winzler, C., Rovere, P., Rescigno, M., Granucci, F., Penna, G., Adorini, L., Zimmermann, V. S., Davoust, J. and Ricciardi-Castagnoli, P. (1997). Maturation stages of mouse dendritic cells in growth factor-dependent long-term cultures. J Exp Med 185(2): 317-328.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Hoffmann, E., Pauwels, A., Alloatti, A., Kotsias, F. and Amigorena, S. (2016). Analysis of Phagosomal Antigen Degradation by Flow Organellocytometry. Bio-protocol 6(22): e2014. DOI: 10.21769/BioProtoc.2014.
Category
Immunology > Immune cell function > Dendritic cell
Cell Biology > Cell-based analysis > Flow cytometry
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












