- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Determination of H2O2 Generation by pHPA Assay
Published: Vol 6, Iss 22, Nov 20, 2016 DOI: 10.21769/BioProtoc.2010 Views: 9077
Reviewed by: Ivan ZanoniAchille BroggiPinchas Tsukerman

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Puromycin Proximity Ligation Assay (Puro-PLA) to Assess Local Translation in Axons From Human Neurons
Raffaella De Pace [...] Saikat Ghosh
Mar 5, 2025 3310 Views

Assay for Site-Specific Homologous Recombination Activity in Adherent Cells, Suspension Cells, and Tumor Tissues
Yuki Yoshino [...] Natsuko Chiba
Apr 5, 2025 2446 Views

Proliferation Assay Using Cryopreserved Porcine Peripheral Mononuclear Cells Stimulated With Concanavalin A and Analyzed With FCS ExpressTM 7.18 Software
Marlene Bravo-Parra [...] Luis G. Giménez-Lirola
Jun 5, 2025 2620 Views
Abstract
The production of reactive oxygen species, including H2O2, is a process that can be used in signaling, cell death, or immune response. To quantify oxidative stress in cells, a fluorescence technique has been modified from a previously described method to measure H2O2 release from cells (Panus et al., 1993; Murthy et al., 2010; Larson-Casey et al., 2016; Larson-Casey et al., 2014; He et al., 2011). This assay takes advantage of H2O2-mediated oxidation of horseradish peroxidase (HRP) to Complex I, which, in turn, oxidizes p-hydroxyphenylacetic acid (pHPA) to a stable, fluorescent pHPA dimer (2,2'-dihydroxy-biphenyl-5,5’ diacetate [(pHPA)2]). The H2O2-dependent HRP-mediated oxidation of pHPA is a sensitive and specific assay for quantifying H2O2 release from cells. This assay can measure H2O2 release from whole cells, mitochondria, or the NADPH oxidase.
Background
H2O2 generation primarily results from dismutation of superoxide anion (O2-), which occurs at a rapid rate (105-106 M-1 s-1) non-enzymatically. Unlike O2-, H2O2 can traverse membranes easily, so it is able to oxidize multiple molecules. ROS can be toxic to cells by oxidizing proteins, lipids, and nucleic acids and are associated with many human diseases. This protocol allows for detection of H2O2 release from NADPH oxidase or mitochondria in various cell types.
Materials and Reagents
- NuncTM 96-well black bottom plate (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 137101 )
- Eppendorf tubes
- 15 and 50 ml conical tubes
- Glucose (Sigma-Aldrich, catalog number: G7528 )
- HEPES (1 M) (Thermo Fisher Scientific, GibcoTM, catalog number: 15630-080 )
- Sodium bicarbonate (NaHCO3) (Thermo Fisher Scientific, Fisher Scientific, catalog number: S233-500 )
- 4-hydroxyphenylacetic acid (pHPA) (Sigma-Aldrich, catalog number: H50004 )
- HRP (Sigma-Aldrich, catalog number: P8125 )
- Ca2+/Mg2+/phenol red-free HBSS (Thermo Fisher Scientific, GibcoTM, catalog number: 14175-095 )
- α-Ketoglutaric acid (Sigma-Aldrich, catalog number: K2000 )
- Hydrogen peroxide solution (H2O2) (30%, w/w) (Sigma-Aldrich, catalog number: H1009 )
- Tris
- EDTA
- Sucrose
- Protease inhibitor tablets (Sigma-Aldrich, catalog number: 11836170001 )
- Phosphatase inhibitors (EMD Millipore, catalog number: 524625 )
- Antimycin A (optional)
- MitoTempo (optional)
- pHPA buffer (see Recipes)
- 1 mM H2O2 working stock solution (see Recipes)
- Mitochondrial buffer (see Recipes)
Equipment
- Kontes pellet pestle motor
- Centrifuge
- Plate reader (Molecular Devices, model: M2 SpectraMax )
Procedure
- Prepare pHPA buffer (see Recipes)
- Prepare H2O2 standard curve (see Figure 1)
Note: Prepared in pHPA buffer prior to the assay (The buffer cannot be stored for subsequent measurements, make fresh at time of use). - 8-12 standards in duplicate or triplicate.
- Load standards into 96-well plate.
Highest concentration of H2O2: 2 µM (2 µl of 1 mM H2O2 working stock solution/ml pHPA buffer)
Serial dilution: 50%
#wells/standard: 2 or 3
µl/well: 200
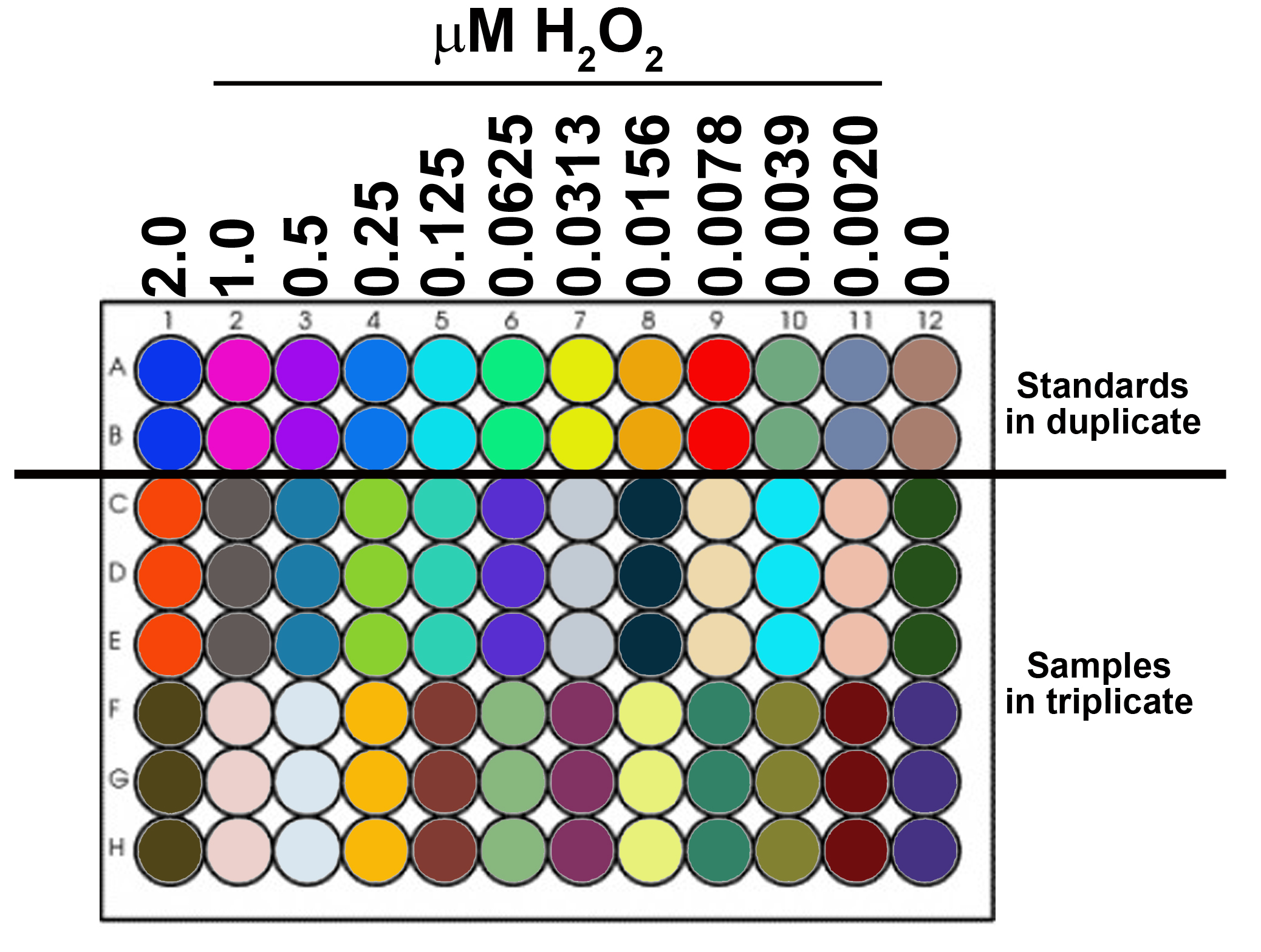
Figure 1. Schematic of 96-well plate with 12 standards (in duplicate) and samples (in triplicate) - Prepare samples
- Sample isolation
- Samples can be whole cells, freshly isolated mitochondria or membrane fractions from various cell types (i.e., macrophages, fibroblasts, bronchoalveolar lavage cells, etc.).
- Mitochondrial isolation:
1) Lyse 10 million macrophages in (100 µl) mitochondrial buffer (see Recipes). Lysis is performed using Kontes Pellet Pestle Motor for 30 sec to homogenize each sample.
2) Centrifuge at 2,000 x g for 10 min at 4 °C, save supernatant at 4 °C.
3) Pellet is lysed and centrifuged [repeat steps 1) and 2)].
4) The two supernatants are pooled and centrifuged at 12,000 x g for 15 min at 4 °C.
5) The pellet is resuspended in (50 µl) mitochondrial buffer without sucrose [same as in 1) but do not add sucrose]. - Each sample should be run in duplicate or triplicate.
- Load samples into 96-well plate:
- Use 5-20 µg protein or 50,000-100,000 cells per well
- Suspend sample in pHPA buffer, mix well
- Load 200 µl/well in duplicate
- Positive controls may be added (i.e., treat cells with 100 μM antimycin A).
- Negative controls may be added (i.e., treat cells with 10 μM MitoTempo).
- Run assay in plate reader
- Pre-warm plate reader to 37 °C.
- Insert plate.
- Run kinetic assay, typically for 4 h with a read every 5-10 min.
- Use an excitation wavelength of 320 nm and emission of 400 nm.
Data analysis
- Use initial readings of standards to generate a standard curve.
- Determine the H2O2 concentration in samples by applying a linear regression to the standard curve and extrapolate.
- Normalize samples to protein or cell number. Samples can be lysed in protein sample buffer to determine cellular protein levels, or alternatively when using live cells, the number of cells can be used to normalize data.
- Data can be expressed as a rate over a set time (Figure 2A) or by showing a kinetic curve (Figure 2B); also see Larson-Casey et al., 2016.
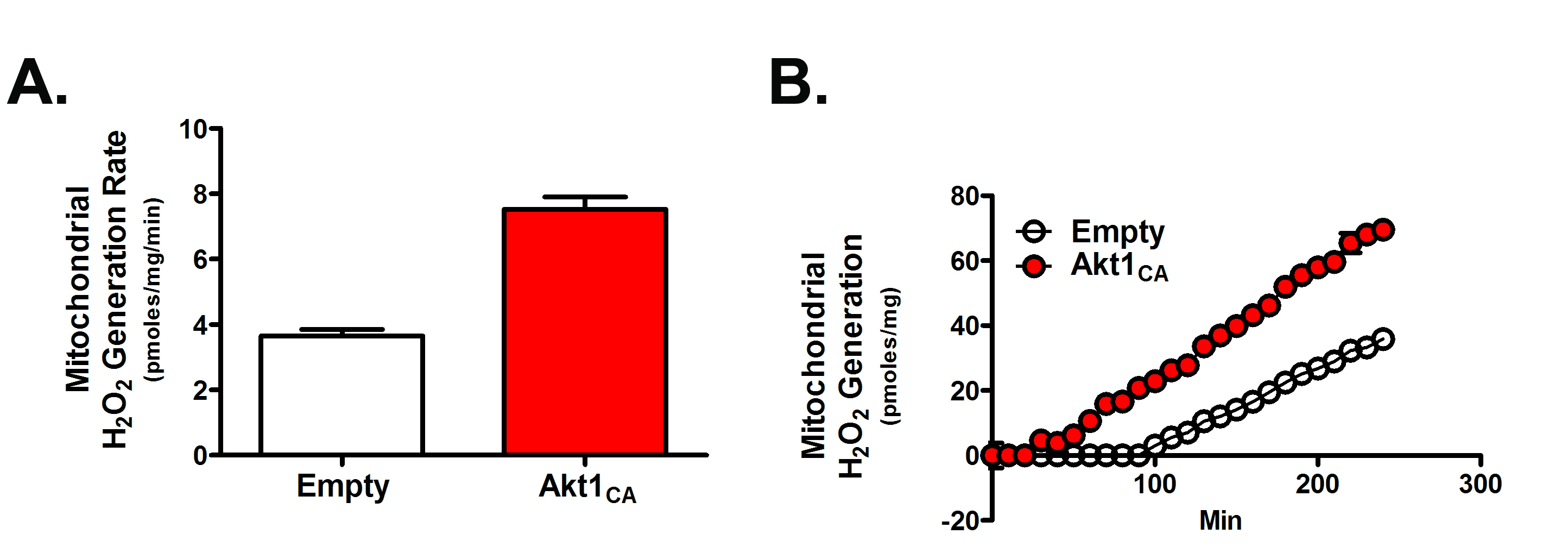
Figure 2. Mitochondrial H2O2 generation in macrophages expressing empty or constitutively active Akt1 (Akt1CA) expression vectors
Notes
- This assay is typically performed on freshly isolated mitochondrial (see Recipes for protocol) or membrane factions.
- Prepare all working solutions on day of use and do not reuse.
Recipes
- pHPA buffer
50 ml total volume made up in Ca2+/Mg2+/phenol red-free HBSS
Make up buffer in 50 ml conical tube and protect from light
- 1 mM H2O2 working stock solution
1.13 µl 30% (w/w) H2O2 stock solution/10 ml H2O
Note: H2O2 MW = 84.01, 30% (w/w) (H2O2 stock solution) equivalent to 8.82 M. - Mitochondrial buffer
10 mM Tris, pH 7.8
0.2 mM EDTA
320 mM sucrose
1 protease inhibitor tablet
Phosphatase inhibitors diluted 1:100
Acknowledgments
This work was supported by 2R01ES015981 & VA merit review BX001135. This protocol was originally adapted from Panus et al. (1993).
References
- He, C., Murthy, S., McCormick, M. L., Spitz, D. R., Ryan, A. J. and Carter, A. B. (2011). Mitochondrial Cu,Zn-superoxide dismutase mediates pulmonary fibrosis by augmenting H2O2 generation. J Biol Chem 286(17): 15597-15607.
- Larson-Casey, J. L., Deshane, J. S., Ryan, A. J., Thannickal, V. J. and Carter, A. B. (2016). Macrophage Akt1 kinase-mediated mitophagy modulates apoptosis resistance and pulmonary fibrosis. Immunity 44(3): 582-596.
- Larson-Casey, J. L., Murthy, S., Ryan, A. J. and Carter, A. B. (2014). Modulation of the mevalonate pathway by Akt regulates macrophage survival and development of pulmonary fibrosis. J Biol Chem 289(52): 36204-36219.
- Murthy, S., Ryan, A., He, C., Mallampalli, R. K. and Carter, A. B. (2010). Rac1-mediated mitochondrial H2O2 generation regulates MMP-9 gene expression in macrophages via inhibition of SP-1 and AP-1. J Biol Chem 285(32): 25062-25073.
- Panus, P. C., Radi, R., Chumley, P. H., Lillard, R. H. and Freeman, B. A. (1993). Detection of H2O2 release from vascular endothelial cells. Free Radic Biol Med 14(2): 217-223.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Larson-Casey, J. L. and Carter, A. B. (2016). Determination of H2O2 Generation by pHPA Assay. Bio-protocol 6(22): e2010. DOI: 10.21769/BioProtoc.2010.
Category
Immunology > Immune cell function > General
Cell Biology > Cell-based analysis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










