- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
PEA-CLARITY: Three Dimensional (3D) Molecular Imaging of Whole Plant Organs
Published: Vol 6, Iss 21, Nov 5, 2016 DOI: 10.21769/BioProtoc.2000 Views: 8957
Reviewed by: Arsalan DaudiTeresa LenserAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Live-Cell Monitoring of Piecemeal Chloroplast Autophagy
Masanori Izumi [...] Shinya Hagihara
Nov 5, 2025 1690 Views

Chloroplast Movement Imaging Under Different Light Regimes With a Hyperspectral Camera
Paweł Hermanowicz [...] Justyna Łabuz
Dec 20, 2025 756 Views

A Simple Protocol for Periodic Live Cell Observation of Flagellate Stages in the Lichen Alga Trebouxia
Enrico Boccato [...] Mauro Tretiach
Jan 20, 2026 186 Views
Abstract
Here we report the adaptation of the CLARITY technique to plant tissues with addition of enzymatic degradation to improve optical clearing and facilitate antibody probe penetration. Plant-Enzyme-Assisted (PEA)-CLARITY, has allowed deep optical visualisation of stains, expressed fluorescent proteins and IgG-antibodies in tobacco and Arabidopsis leaves. Enzyme treatment enabled penetration of antibodies into whole tissues without the need for any sectioning of the material. Therefore, this protocol facilitates protein localisation of intact tissue in 3D whilst retaining cellular structure.
Background
Fixation and embedding of plant tissue for molecular interrogation using techniques such as histological staining, immunohistochemistry or in situ hybridisation has been the foundation of cell biology studies for decades. Applying these techniques for 3D tissue analysis is seriously limited by the need to section the tissue, image each section, and then reassemble the images into a 3D representation of the structures of interest. Here we present a fundamental shift from the two dimensional plane to that of three dimensions whilst retaining molecular structures of interest without the need to section the plant tissue. Recent advances in fixation and ‘clearing’ techniques such as SeeDB, ScaleA2, 3DISCO, CLARITY and its recent variant PACT enabled intact imaging of whole embryos, brains and other organs in mouse and rat models. The new CLARITY system fixes and binds tissues within an acrylamide mesh structure. Proteins and nucleic acids are covalently linked to the acrylamide mesh by formaldehyde, then optically interfering lipid structures of animal cell membranes are removed using detergent (SDS). This renders such tissue optically transparent and suitable for deep imaging of up to ~5 mm using confocal microscopy.
Materials and Reagents
- 50 ml conical tube
- 1.5 ml microfuge tubes
- Aluminum foil
- Parafilm (Sigma-Aldrich, catalog number: P-7793 )
- Lint free paper
- 1.5 ml Protein LoBind tubes (Eppendorf, catalog number: 0030108116 )
- Glass microscope slide
- Glass microscope coverslip
- Nicotiana tabacum (Hanson and Köhler, 2001)
- 16% paraformaldehyde (Electron Microscopy Sciences, catalog number: 15710 )
- Sodium azide (NaN3) (Sigma-Aldrich, catalog number: S-2002 )
- 0.005% NaN3 in PBS (N3PBS)
- Triton X-100 (Sigma-Aldrich, catalog number: T-9284 )
- Dulbecco's phosphate buffered saline (DPBS, autoclaved) (Thermo Fisher Scientific, GibcoTM, 21600-010 )
- 0.1% Triton X-100 in PBS (PBST)
- Vaseline (Unilever, VASELINE®)
- BluTack (Bostic)
- Rubisco antibody (rabbit) (Gift - Spencer Whitney, Whitney and Andrews, 2001)
- Cy5 secondary AB (anti-rab) (Abcam, catalog number: Ab6564 )
- Propidium iodide (Sigma-Aldrich, catalog number: P-4864 )
- Calcofluor white (Sigma-Aldrich, catalog number: F-3543 )
- 40% acrylamide (Bio-Rad Laboratories, catalog number: 161-0140 )
- 2% bis acrylamide (Bio-Rad Laboratories, catalog number: 161-0142 )
- VA-044 initiator (Wako Pure Chemical Industries, catalog number: 017-19362 )
- Deionized and distilled water (ddH2O)
- Sodium dodecyl sulfate (SDS) (Sigma-Aldrich, catalog number: L-3771 )
- Boric acid (H3BO3) (Sigma-Aldrich, catalog number: B-6768 )
- Sodium hydroxide (NaOH) (Sigma-Aldrich, catalog number: S-8045 )
- α-amylase (Megazyme, catalog number: E-ANAAM )
- α-L-arabinofuranosidase (Megazyme, catalog number: E-ABFCJ )
- β-mannanase (Megazyme, catalog number: E-BMACJ )
- Cellulase (Megazyme, catalog number: E-CELBA )
- Pectate lyase (Megazyme, catalog number: E-PLYCJ )
- Xyloglucanase (Megazyme, catalog number: E-XEGP )
- Calcium chloride (CaCl2) (Sigma-Aldrich, catalog number: C-5670 )
- Hydrogel solution (200 ml) (see Recipes)
- SDS clearing solution (1 L) (see Recipes)
- Enzyme treatment solution (10 ml) (see Recipes)
Notes:
- Please refer to MSDS before conducting protocol as paraformaldehyde (PFA), acrylamide, sodium dodecyl sulfate (SDS) and sodium azide (NaN3) are known irritants, sensitizers, carcinogens and neurotoxins. The use of personal protective equipment (PPE) is imperative whilst undertaking this protocol.
- Any specific IgG primary antibody and respective secondary antibody can be used with this protocol.
- Protocol can be paused and samples stored at any stage from step D onwards in either SDS clearing solution or N3PBS.
Equipment
- Vacuum pump at -100 kPa
- Fume hood
- 4 °C fridge
- 37 °C water bath
- Weigh balance
- 37 °C incubator shaker
- Leica SP8 confocal microscope/lightsheet microscope or equivalent
- Long working distance objectives greater than 2 mm
Software
- Leica Applications Suite - Fluorescence (LAS-AF) software
Procedure
- Plant harvesting
- Harvest mature, fully expanded leaves at the end of the dark period to minimise starch accumulation. Replication and position within the leaf of excised disk will differ depending on experimental design.
- Harvest mature, fully expanded leaves at the end of the dark period to minimise starch accumulation. Replication and position within the leaf of excised disk will differ depending on experimental design.
- Fixation
- Excise ~20 N. tabacum 7 mm leaf disks of each line from fully expanded leaves before immediately placing each sample into a single 50 ml conical tube containing ice cold hydrogel solution. If samples are required to be individually separated, use 1.5 ml tubes for each individual sample.
- Place tissue under vacuum at -100 kPa (in fume hood) for 1-2 h on ice and in darkness if fluorophores are present to facilitate infiltration of hydrogel and removal of gas.
- Transfer to a 4 °C fridge overnight.
Note: If fluorophores are present, keep samples in darkness throughout protocol, i.e., wrap tube(s) in aluminium foil.
- Excise ~20 N. tabacum 7 mm leaf disks of each line from fully expanded leaves before immediately placing each sample into a single 50 ml conical tube containing ice cold hydrogel solution. If samples are required to be individually separated, use 1.5 ml tubes for each individual sample.
- Hydrogel polymerization (Figure 1.1)
- Carefully remove individual leaf disks from the 50 ml conical tube and place a single disk into 1.5 ml microfuge tubes containing 1 ml fresh chilled hydrogel solution and keep on ice.
- Place samples under vacuum for 15 min to remove excess gas from the transferring of samples.
- Completely fill 1.5 ml tubes with hydrogel solution taking care to remove any air bubbles before sealing with Parafilm.
- Float sealed tubes in a 37 °C water bath overnight to polymerize.
- Carefully remove individual leaf disks from the 50 ml conical tube and place a single disk into 1.5 ml microfuge tubes containing 1 ml fresh chilled hydrogel solution and keep on ice.
- Tissue clearing (Figure 1.2)
- Remove polymerized leaf disks from 1.5 ml tubes and separate excess hydrogel carefully from the sample with lint free paper (see Note below).
- Wash samples in separate 50 ml conical tubes of SDS clearing solution and change 3 times daily for 2 days to remove excess unbound PFA and acrylamide at room temperature. Take care to dispose of this solution correctly.
- 50 ml of SDS clearing solution was replaced daily in each tube for a period of 4-6 weeks (or until clear) at 37 °C with very gentle agitation.
Note: Peel away excess hydrogel by placing sample onto clean area of lint free paper and pull apart slowly. Repeat this process until all excess hydrogel is removed from the surface of the leaf disk. The sample is quite robust at this stage and even perpendicular trichome parturitions will remain after removal of excess hydrogel if completed with care.
- Remove polymerized leaf disks from 1.5 ml tubes and separate excess hydrogel carefully from the sample with lint free paper (see Note below).
- Enzyme treatment (Figure 1.3)
- Extensively wash samples in 50 ml N3PBS (see Materials and Reagents section) with 3 changes each day for 3 days at room temperature.
Note: SDS is an inhibitor of enzymatic activity and improper washing will result in incomplete enzymatic degradation of the cell wall. - Transfer individual sample to a 1.5 ml Protein LoBind tube containing 1 ml of enzyme mix ([EM], see Materials and Reagents section) and keep at 37 °C for 5-7 days with very gentle agitation.
- During enzyme treatment a vacuum of -100 kPa is applied in 3 x 5 min bursts, 3 x daily to help facilitate enzyme infiltration.
- Replace solution with fresh EM on the third day.
- During enzyme treatment a vacuum of -100 kPa is applied in 3 x 5 min bursts, 3 x daily to help facilitate enzyme infiltration.
- Carefully remove samples after 5-7 days then wash in 50 ml PBST with three changes over a 24 h period with gentle agitation.
- Extensively wash samples in 50 ml N3PBS (see Materials and Reagents section) with 3 changes each day for 3 days at room temperature.
- Immunolocalization
- Dilute primary/secondary antibodies to desired concentration in PBST.
- Transfer sample into 1.5 ml Protein LoBind tube containing 1 ml desired primary antibody/s concentration at 37 °C for 5 days with very gentle agitation.
- During antibody treatment a vacuum of -100 kPa is applied in 3 x 5 min bursts, 3 x daily to help facilitate primary antibody infiltration.
- Transfer sample into 1.5 ml Protein LoBind tube containing 1 ml desired primary antibody/s concentration at 37 °C for 5 days with very gentle agitation.
- Wash 3 times with 50 ml PBST for 24 h period.
- Follow steps E2a and E2b above for secondary antibody/s.
- Wash 3 times with 50 ml N3PBS for a 24 h period.
Note: For multiple rounds of immunohistochemistry using the same sample, repeat protocol steps D1 and D2 to strip previous antibody after imaging then re-probe using a different set of primary and secondary antibodies as outlined on protocol steps F1-F4.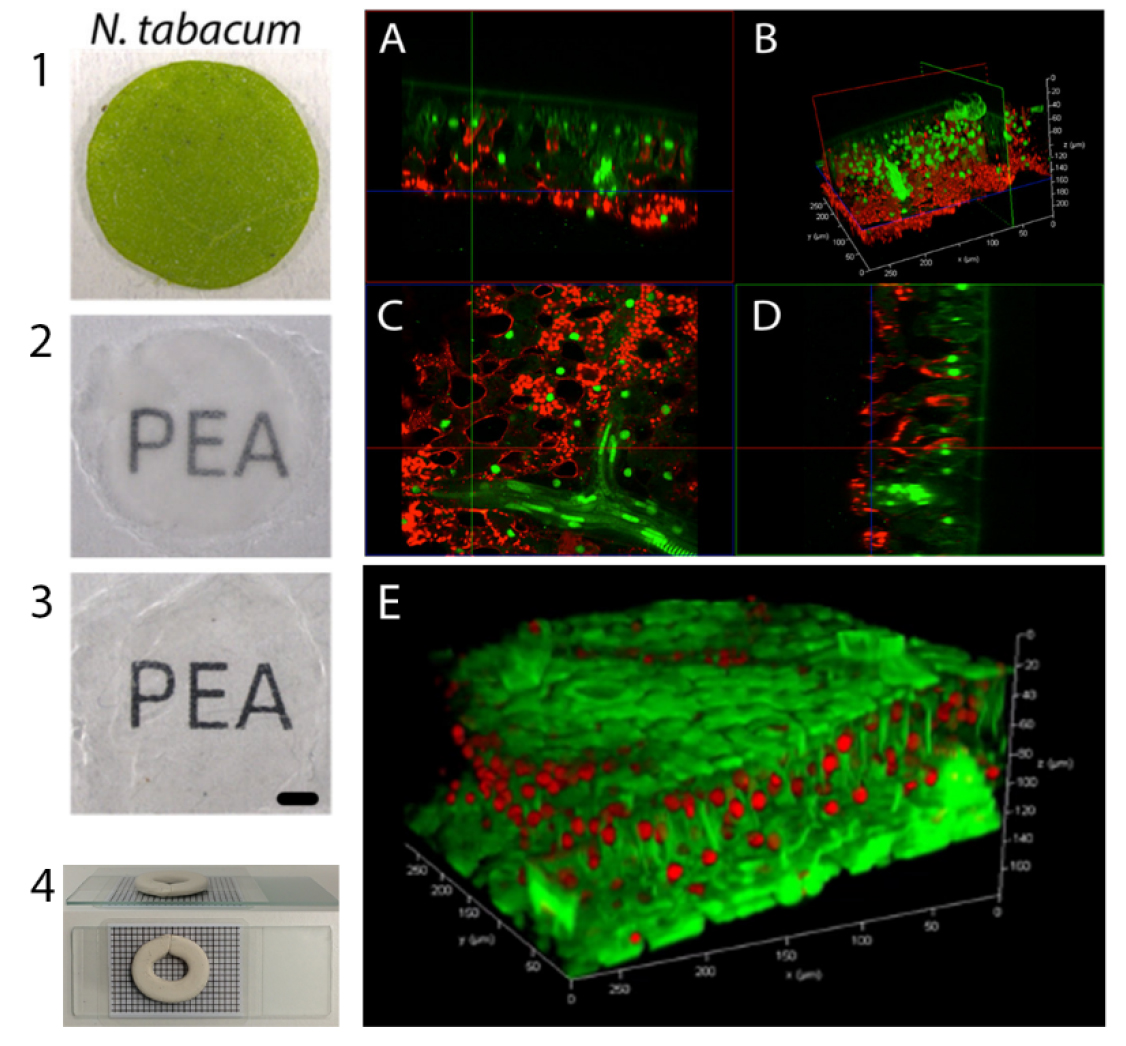
Figure 1. Clearing, mounting and imaging of N. tabacum leaf. Left panel: 1. Fresh leaf disc from a fully expanded N. tabacum leaf. 2. Fixed, hydrogel embedded, passively cleared leaf disk. 3. Cleared cell wall enzyme treated leaf disk for immunohistochemistry and imaging. 4. Example of mounting procedure for confocal imaging. Scale bar = 1 mm. Right panel: A. CLSM 3D projection of a passively cleared, cell wall enzyme treated (PEA-CLARITY) Sv-40 (nuclear localised GFP-green) N. tabacum leaf, immunostained with tobacco RuBisCO primary and Cy5 secondary antibodies (red). The 3D projection is shown in (B) and the x, y, z slices are shown in (A, C, D) respectively. E. CLSM 3D projection of a passively cleared (without cell wall enzyme digestion) N. tabacum leaf showing nuclei stained with propidium iodide (red), and cell walls stained with calcofluor white (green). The 3D projection was generated with Leica Applications Suite - Fluorescence (LAS-AF) software. See, Palmer et al. (2015) for further details.
- Dilute primary/secondary antibodies to desired concentration in PBST.
- Imaging preparation (Figure 1.4)
Note: During this protocol samples were mounted in PBS however, as described in (Palmer et al., 2015) other mounting mediums such as Focus Clear (Chung et al., 2013) and RIMS (Yang et al., 2014) can be used to optically match the hydrogel to enhance image clarity. If using other mounting mediums, then samples will need to be incubated prior to mounting.- Using a small piece of BluTack create a well by rolling the putty into a long cylindrical shape and apply to a glass microscope slide so the sides of the well are just higher than the sample thickness (~2 mm).
- Seal the outer rim of the BluTack well and the microscope slide with a thin layer of Vaseline.
- Half fill well with PBS (or mounting medium).
- Place sample into well and cover with PBS (or mounting medium).
- Place a glass microscope coverslip over the well ensuring there are no air bubbles.
- Samples are now ready for imaging.
- Using a small piece of BluTack create a well by rolling the putty into a long cylindrical shape and apply to a glass microscope slide so the sides of the well are just higher than the sample thickness (~2 mm).
Data analysis
All images displayed in this article are raw images taken from the Leica SP8 (Figures 1A-1E). Unprocessed 3D reconstructions were performed in the Leica Applications Suite - Fluorescence (LAS-AF) software.
Recipes
- Hydrogel solution (200 ml)
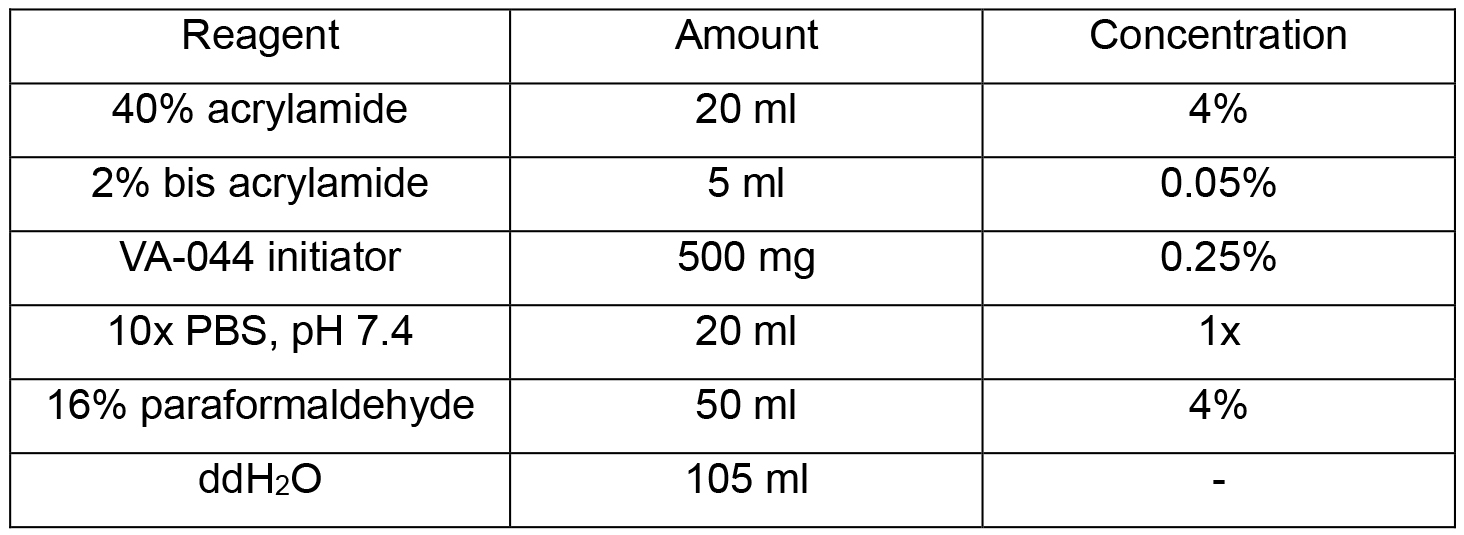
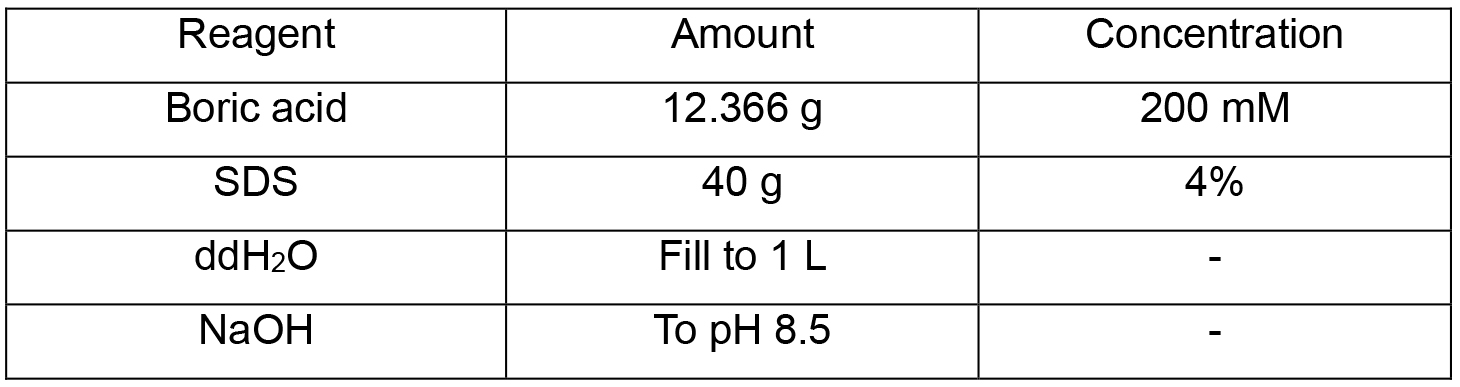
Note: Store at 4 °C. - SDS clearing solution (1 L)
- Enzyme treatment solution [EM] (10 ml)
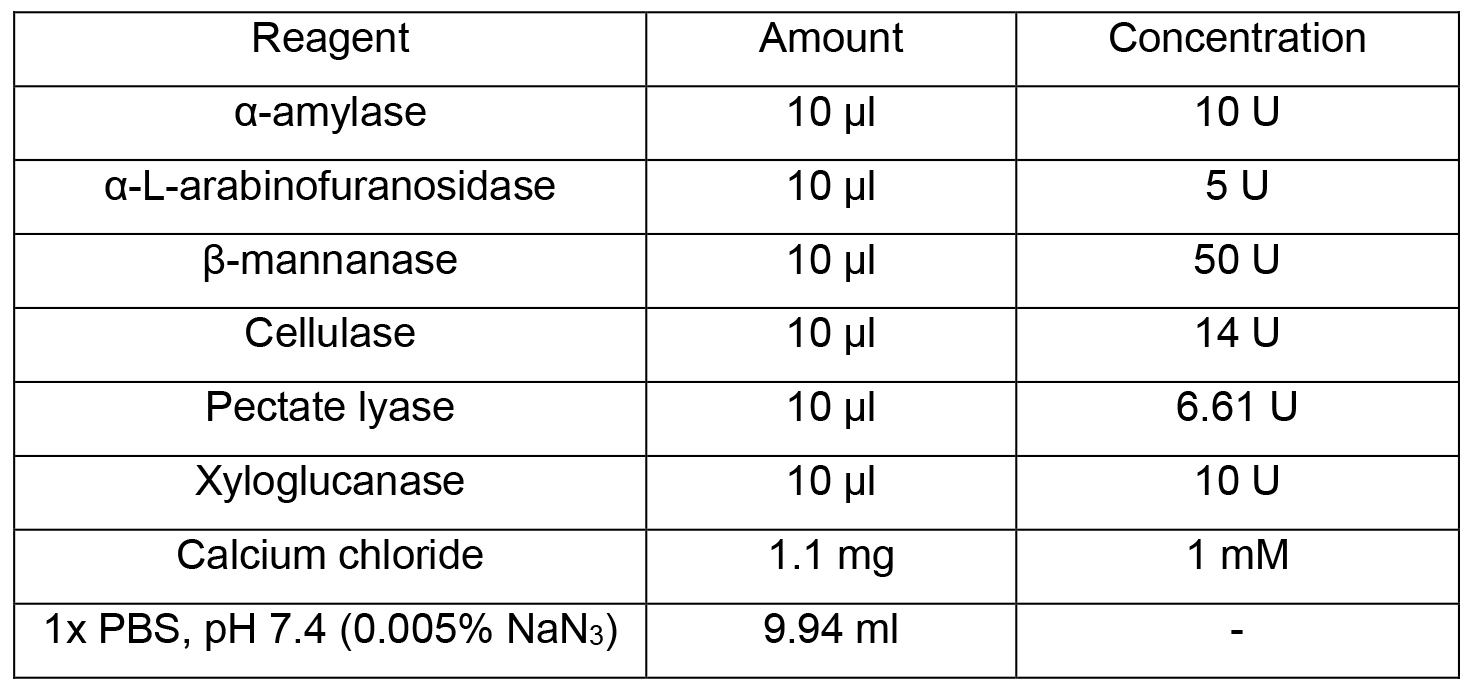
Note: Make fresh, do not store. Only use enzymes that have passed a size separation gel quality control procedure to assess purity such as those listed from Megazyme.
Acknowledgments
We would like to thank Mark Talbot for helpful suggestions during CLSM measurements, David McCurdy for providing the Sv-40 N. tabacum line, Spencer Whitney for providing the RuBisCO antibody, Vivien Rolland for initial CLSM investigations, and Joe Enright for growing the plants.
References
- Chung, K., Wallace, J., Kim, S. Y., Kalyanasundaram, S., Andalman, A. S., Davidson, T. J., Mirzabekov, J. J., Zalocusky, K. A., Mattis, J., Denisin, A. K., Pak, S., Bernstein, H., Ramakrishnan, C., Grosenick, L., Gradinaru, V. and Deisseroth, K. (2013). Structural and molecular interrogation of intact biological systems. Nature 497(7449): 332-337.
- Hanson, M. R. and Kohler, R. H. (2001). GFP imaging: methodology and application to investigate cellular compartmentation in plants. J Exp Bot 52(356): 529-539.
- Palmer, W. M., Martin, A. P., Flynn, J. R., Reed, S. L., White, R. G., Furbank, R. T. and Grof, C. P. (2015). PEA-CLARITY: 3D molecular imaging of whole plant organs. Sci Rep 5: 13492.
- Whitney, S. M. and Andrews, T. J. (2001). The gene for the ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) small subunit relocated to the plastid genome of tobacco directs the synthesis of small subunits that assemble into Rubisco. Plant Cell 13(1): 193-205.
- Yang, B., Treweek, J. B., Kulkarni, R. P., Deverman, B. E., Chen, C. K., Lubeck, E., Shah, S., Cai, L. and Gradinaru, V. (2014). Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 158(4): 945-958.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Palmer, W. M., Martin, A. P., Flynn, J. R., Reed, S., White, R., Furbank, R. T. and Grof, C. P. L. (2016). PEA-CLARITY: Three Dimensional (3D) Molecular Imaging of Whole Plant Organs. Bio-protocol 6(21): e2000. DOI: 10.21769/BioProtoc.2000.
Category
Plant Science > Plant cell biology > Cell imaging
Plant Science > Plant physiology > Phenotyping
Cell Biology > Cell imaging > Live-cell imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









