- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Determination of the Glycolysis and Lipogenesis in Culture of Hepatocytes
Published: Vol 6, Iss 21, Nov 5, 2016 DOI: 10.21769/BioProtoc.1993 Views: 12867
Reviewed by: Jia LiXuecai GeXiujun Fan

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Intestinal Co-culture System to Study TGR5 Agonism and Gut Restriction
Snehal N. Chaudhari and A. Sloan Devlin
Mar 20, 2021 6658 Views

OrganoPlate Micro-fluidic Microvessel Culture and Analysis
Abidemi Junaid and Thomas Hankemeier
Jul 5, 2021 4543 Views

Mass Spectrometry-based Lipidomics, Lipid Bioenergetics, and Web Tool for Lipid Profiling and Quantification in Human Cells
Liang Cui [...] Kuan Rong Chan
Aug 20, 2023 3017 Views
Abstract
Metabolic flux analyses are needed to provide insights into metabolic regulation that occurs in cells. The current protocol describes fast and reproducible methods for determining glycolysis and de novo lipogenesis of hepatocytes. Primary culture of hepatocytes is an ‘in vitro’ model useful to study liver glucose and lipid metabolism (Denechaud et al., 2016). The protocol is divided in 2 parts. Part I: Glycolysis experiment is assessed using the Seahorse extracellular flux (XF) analyser. Glycolysis is determined via the measurement of the extracellular acidification rate (ECAR) of the media, which come predominately from the cellular excretion of lactic acid after the conversion of glucose in pyruvate. Part II: De novo lipogenesis experiment determines the radioactive C14 incorporation in triglycerides (TG) from acetate C14 precursor. After 2 h acetate supplementation to the media lipids are extracted and separated by TLC (Thin Layer Chromatography) prior quantification of newly synthetized TG labelled.
Background
There are different approaches for evaluating glucose and lipid metabolism: metabolite quantification, enzyme activity, and metabolomics... Our protocols focus on metabolic flux analyses of live cells and do not need a metabolomic facility. The Seahorse extracellular flux (XF) analyzer, which is now present in a lot of institution, is a powerful tool for measuring indirectly glycolysis in live cells by determining media pH. Lipogenesis protocol does not need a big investment and is highly reproducible. It could also be determined using tritiated water, which is incorporated by the Fatty Acid Synthase in de novo synthetized lipids.
Materials and Reagents
- 60 mm tissue culture dishes (TPP, catalog number: 93060 )
- XF assay 24 well cartridge and cell culture microplate V7 (Agilent Technologies, Seahorse Bioscience, catalog number: 100850-001 )
- 8-15 weeks old C57BL/6 mouse (Janvier lab)
- Collagen type I, rat tail (EMD Millipore, catalog number: 08-115 )
- M199 medium (Thermo Fisher Scientific, GibcoTM, catalog number: 41150020 )
- Trypsin (2.5%), no phenol red (Thermo Fisher Scientific, GibcoTM, catalog number: 15090046 )
- Fetal bovine serum (FBS)
- DMEM without glucose, L-glutamine, phenol red, sodium pyruvate and sodium bicarbonate, powder, suitable for cell culture (Sigma-Aldrich, catalog number: D5030-10X1L )
- D-(+)-glucose solution (Sigma-Aldrich, catalog number: G8769 )
- L-glutamine, 200 mM (Thermo Fisher Scientific, GibcoTM, catalog number: 25030081 )
- XF calibrant solution (Agilent Technologies, Seahorse Bioscience, catalog number: 100840-000 )
- PierceTM BCA Protein Assay Kit (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 23225 )
- Sodium chloride (NaCl) (AppliChem, catalog number: A3597 )
- Phenol red sodium salt (Sigma-Aldrich, catalog number: P5530 )
- Tris, pH 7.5 (Applichem, catalog number: A1379 )
- EDTA (Sigma-Aldrich, catalog number: E6758 )
- Triton X-100 (Sigma-Aldrich, catalog number: X100 )
- Protease inhibitor (Sigma-Aldrich, catalog number: P8340 )
- Glycolysis media (see Recipes)
- 9x glucose solution (see Recipes)
- Lysis Buffer (see Recipes)
Equipment
- CO2-free incubator set to 37 °C (Memmert or other suppliers)
- 5% CO2 incubator set to 37 °C (SalvisLab or other suppliers)
- Centrifuge
- XF 24 extracellular flux analyser (Seahorse Bioscience)
- Cell culture hood (Vitaris or other suppliers)
- pH meter (Mettler Toledo or other suppliers)
- Cell counter (Thermo Fisher Scientific or other suppliers)
Software
- Seahorse software (XFReader 24 version 1.6 supply with the XF 24 extracellular flux analyser)
Procedure
- Isolate hepatocytes from 8-15 week-old C57BL6/J mice according to Dentin et al. (2004). Dilute at 50 µg/ml the stock collagen I in PBS (do not re-use the solution because collagen I will precipitate at neutral pH). Coat 60 mm culture dish with collagen I (10-20 min at room temperature [RT] are sufficient). Then plate 2 million hepatocytes per dish and incubate in 5% CO2 incubator at 37 °C. 2 h later, change the medium with fresh M199.
- At least 6 h before beginning the glycolysis experiment soak the cartridge in a CO2-free incubator at 37 °C with 1 ml of XF calibrant per well.
- 24 h after hepatocytes isolation, coat the 24 well of Seahorse XF 24 plate with collagen I (50 µg/ml, 10 min at RT). Then detach the cells by incubating with trypsin for 1-2 min. Stop trypsin with 10% FBS supplemented media. Centrifuge and count the hepatocytes with automatic cell counter. Plate 40,000 cells per well in 100 µl of glycolysis media. Keep 4 wells with only 100 µl of media (without cells) for the background correction of XF24 extracellular flux analyser.
Note: After seeding, hepatocytes viability should be more than 80%. - Incubate cells 30 min at 37 °C in CO2-free incubator as indicated by the manufacturer (CO2 could participate to the acidification of the media by its hydration to carbonic acid and bicarbonate). Then add 500 µl of 37 °C glycolysis media in each well (volume total of 600 µl).
- During these 30 min, load the port A of the cartridge (Figure 1) containing the probes with 75 µl of 9x glucose solution. Prepare the protocol on the Seahorse software according to manufacturer. Choose 3 min mix, 2 min wait and 3 min measurement for the parameters of the Seahorse assay.
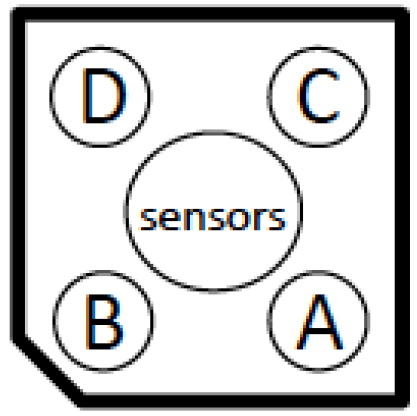
Figure 1. Injection port layout - Start the protocol and load the cartridge with the calibrant to the XF24 Seahorse apparatus (follow the software indications). At the end of the calibration, replace the calibrant plate by the plate with the cells.
- After the first measurement, in function of hepatocytes respiration, modify the program: if OCR (oxygen consumption rate) is higher than 200 pMoles oxygen/min, change the program to 4 min mix, 2 min wait and 2 min measurement (according to manufacturer instructions).
Note: For hepatocytes respiration, OCR should not be below 100 pMoles oxygen/min. - 2 or 3 measurements under basal and 10 measurements after glucose injection are recommended.
- At the end of the run, lyse the cells in 40 µl of lysis buffer on ice for 30 min. Quantify the protein level using BCA protein assay.
Data analysis
- The results are normalized by the protein content using the Seahorse software according to the manufacturer and can be represented as Figure 2.
- Experiment must be done at least 2 times with 8 replicates.
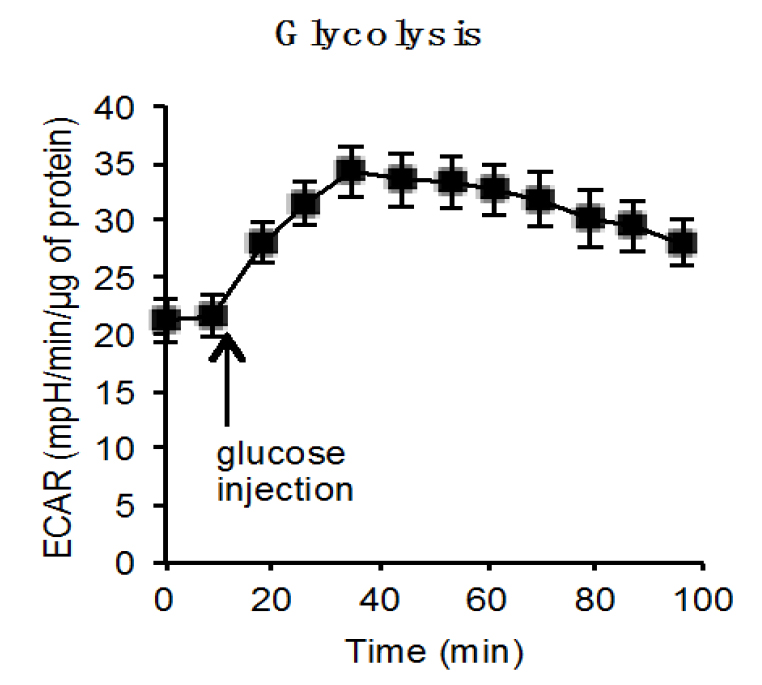
Figure 2. Representative ECAR plot with injection of glucose. ECAR of C57Bl6/J hepatocytes before and after glucose injection using Seahorse analyser (2 independent experiments, each with 8 replicates).
Notes
- The protocol can be adapted to specific needs (use of genetically modified hepatocytes, other cell types).
Recipes
- Glycolysis media
DMEM without glucose, L-glutamine, phenol red, sodium pyruvate and sodium bicarbonate, powder, suitable for cell culture
143 mM NaCl
3 mg/L phenol red
2 mM glutamine
Adjust pH to 7.35 at 37 °C - 9x glucose solution (225 mM)
4.55 ml glycolysis media
450 µl 2.5 M glucose
Adjust pH to 7.35 at 37 °C - Lysis buffer
150 mM NaCl
50 mM Tris (pH 7.5)
5 mM EDTA
1% Triton X-100
Protease inhibitor (1x)
Materials and Reagents
- 6 wells plate (Corning, Costar®, catalog number: 3335 )
- Scrapers (Corning, catalog number: 3010 )
- Glass Pasteur pipet
- Plastic bag
- 8-15 weeks old C57BL/6 mouse (Janvier lab)
- Nitrogen gas (Carbagaz or other suppliers)
- Collagen type I, rat tail (EMD Millipore, catalog number: 08-115 )
- M199 medium (Thermo Fisher Scientific, GibcoTM, catalog number: 41150020 )
- Dexamethasone (Sigma-Aldrich, catalog number: D1756 )
- Insulin (Actrapid HM, Novo Nordisk)
- Phosphate-buffered saline (PBS)
- D-(+)-glucose solution (Sigma-Aldrich, catalog number: G8769 )
- Acetic acid, sodium salt, [1-14C], 1 µCi/µl (PerkinElmer, catalog number: NEC084H001MC )
- Chloroform (Sigma-Aldrich, catalog number: 34854 )
- Methanol (Sigma-Aldrich, catalog number: 34860 )
- Pre-coated TLC-sheets, 0.2 mm silica gel (MACHEREY-NAGEL, catalog number: 805013 )
- Iodine (Sigma-Aldrich, catalog number: 207772 )
- Diethyl ether (Sigma-Aldrich, catalog number: 309966 )
- Petroleum ether (Sigma-Aldrich, catalog number: 320447 )
- Acetic acid (Sigma-Aldrich, catalog number: 695092 )
- Chloroform/methanol (2:1) (see Recipes)
- TLC migration solvent (see Recipes)
Equipment
- 5% CO2 incubator set to 37 °C (SalvisLab or other suppliers)
- Cell culture hood (Vitaris or other suppliers)
- Sonication water bath (Bioruptor)
- Vortex (Scientific Industries or other suppliers)
- TLC developing chamber for 20 x 20 cm plates (with lid) (CAMAG, catalog number: 022.5250 )
- Chemical hood
- Cyclone Plus Storage Phosphor Scanner (PerkinElmer, catalog number: C431200 )
- MS Multisensitive Phosphor Screens (PerkinElmer, catalog number: 7001723 )
- Centrifuge
Software
- OptiQuant software (supply with the Cyclone Plus Storage Phosphor Scanner)
Procedure
- Isolate hepatocytes from 8-15 week-old C57BL6/J mice according to Dentin et al. (2004). As previously described, coat the wells of a 6 wells-plate with collagen I (10 min at RT) and plate 1 million cells per well. After 2 h, change the medium with fresh M199.
- The next day, treat the cells with the non-stimulated (G5) and stimulated (G25i) conditions for 24 h:
a.Low-glucose (G5): M199 media (glucose 5 mM) plus dexamethasone 10-7 M.
b.High-glucose insulin (G25i): M199 media (glucose 5 mM) complemented to 25 mM glucose, plus insulin 100 nM and dexamethasone 10-7 M. - Add acetate C14 (0.5 µCi per well [0.5 µl]) in the media. C14 will be incorporated in de novo synthetized lipids.
- After 2 h, wash the cells 2 times with PBS and scrap the cells of each well in 400 µl of PBS. Then the 400 µl is divided into 100 µl and 300 µl for protein quantification and lipid extraction respectively as described below.
- For protein quantification. Freeze at -20 °C to improve the lysis of the protein. (Optional: sonicate 15 sec in sonication water bath). Quantify using BCA protein assay.
- From the 300 µl left, perform lipid extraction under a chemical hood as follow. Add 100 µl of M199 media with phenol red to facilitate the recognition of the 2 phases in the later stage. Then add 1.6 ml of chloroform/methanol (2:1) and vortex 5 to 10 min. Centrifuge 5 min at 15,000 x g (RCF) at RT and collect the organic phase (lower phase) with glass Pasteur pipet. The phenol red helps to distinguish two phases. Then evaporate under nitrogen blowing (approx. 20 to 40 min) and resuspend in 150 µl of chloroform/methanol (2:1).
- Add 1 cm of TLC migration solvent in the TLC developing chamber 20 min before loading the TLC plate in the TLC developing chamber.
- Mark a line with a pencil at 2 cm from the bottom of the TLC plate. Samples will be loaded every 1.5-2 cm on this line. Load 40 µl as a dot on the TLC plate. To avoid too large dots, at the same time you load, blow nitrogen for immediate drying.
- Load the TLC plate in the TLC developing chamber. Allow TLC plate migration to 1 cm of the top (approx. 45 min).
- (Optional) After TLC migration, the lipid can be visualised. Under the chemical hood, dry the TLC plate under the hood and colour lipid with iodine in a clean TLC chamber. Add some iodine stones in the bottom of the TLC chamber and place the TLC plate. Wait until the brown colour appears (10 to 20 min). You can take a picture under the hood or you can put the plate in a closed plastic bag for scanning. Recover the iodine stones in a close bottle (they can be used indefinitely). Note that the coloration disappears outside of the chamber. Remove the coloration under the hood (approx. 10 min).
- Expose the TLC to the MS Multisensitive Phosphor Screens for at least 5 days in a film cassette. Then scan the screen using Cyclone Plus Storage Phosphor Scanner (Figure 3).
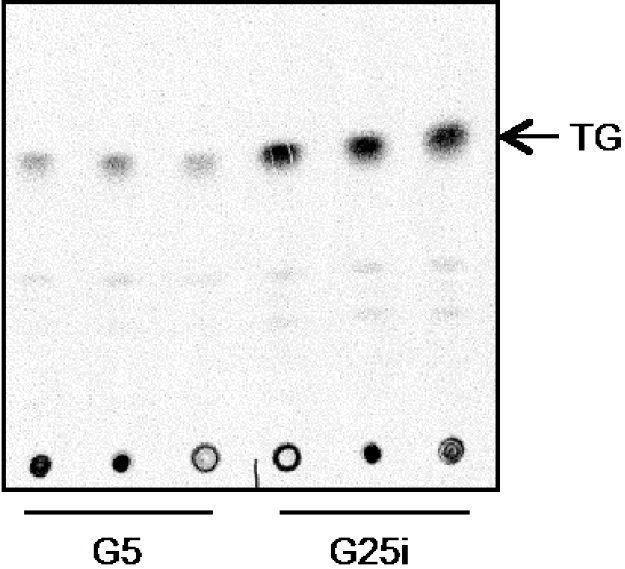
Figure 3. Representative scan after migration of the sample on TLC plate. Visualization of C14 labelled lipids after TLC migration. Samples from hepatocytes treated 24 h with G5 or G25i where load on the bottom of the plate. TGs are highlighted.
Data analysis
- Using the OptiQuant software supplied with the Scanner, determine the levels of incorporated C14 in the TG by quantifying the TG spot intensity.
- The results are normalized by the protein content and expressed as relative values respect to the control condition (G25i) taken as 100% of incorporation.
- Experiments must be performed 3 times in triplicate, as minimum.
Notes
- Each condition has to be performed in triplicate. G5 and G25i are conditions were lipogenesis is low and high respectively in wild type hepatocytes. Hepatocytes in G25i are expected to have 3 times more lipogenesis (Denechaud et al., 2016).
- Lipid extraction, TLC migration, TLC coloration (iodine) have to be performed under a chemical hood.
- Any phosphorimager scanner can be used for scanning the TLC plate.
- The protocol can be adapted to specific needs (specific culture conditions, use of genetically modified hepatocytes, other cell types).
Recipes
- Chloroform/methanol (2:1)
2 volumes of chloroform for 1 volume of methanol - TLC migration solvent
127 ml petroleum ether
22 ml diethyl ether
0.5 ml acetic acid
Acknowledgments
We thank Dr. Isalel Lopez-Mejia for helpful discussions and for her expertise in XF24 extracellular flux analyser. This work was supported by grants from the Swiss Ligue Contre le Cancer, the Swiss National Science Foundation and the Fondation de France. We thank Dr. K. Schooonjans (EPFL, Lausanne, Switzerland) for his previous work (Oosterveer et al., 2012) that we adapt and modify the glycolysis protocol.
References
- Dentin, R., Pegorier, J. P., Benhamed, F., Foufelle, F., Ferre, P., Fauveau, V., Magnuson, M. A., Girard, J. and Postic, C. (2004). Hepatic glucokinase is required for the synergistic action of ChREBP and SREBP-1c on glycolytic and lipogenic gene expression. J Biol Chem 279(19): 20314-20326.
- Denechaud, P. D., Lopez-Mejia, I. C., Giralt, A., Lai, Q., Blanchet, E., Delacuisine, B., Nicolay, B. N., Dyson, N. J., Bonner, C., Pattou, F., Annicotte, J. S. and Fajas, L. (2016). E2F1 mediates sustained lipogenesis and contributes to hepatic steatosis. J Clin Invest 126(1): 137-150.
- Oosterveer, M. H., Mataki, C., Yamamoto, H., Harach, T., Moullan, N., van Dijk, T. H., Ayuso, E., Bosch, F., Postic, C., Groen, A. K., Auwerx, J. and Schoonjans, K. (2012). LRH-1-dependent glucose sensing determines intermediary metabolism in liver. J Clin Invest 122(8): 2817-2826.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Denechaud, P. and Fajas, L. (2016). Determination of the Glycolysis and Lipogenesis in Culture of Hepatocytes. Bio-protocol 6(21): e1993. DOI: 10.21769/BioProtoc.1993.
Category
Cell Biology > Cell metabolism > Lipid
Cell Biology > Cell metabolism > Carbohydrate
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









