- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Rapid Determination of Cellulose, Neutral Sugars, and Uronic Acids from Plant Cell Walls by One-step Two-step Hydrolysis and HPAEC-PAD
(*contributed equally to this work) Published: Vol 6, Iss 20, Oct 20, 2016 DOI: 10.21769/BioProtoc.1978 Views: 18451
Reviewed by: Renate WeizbauerYingnan HouAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Enzymatic Starch Quantification in Developing Flower Primordia of Sweet Cherry
Nestor Santolaria [...] Afif Hedhly
Apr 5, 2025 1896 Views

New Approach to Detect and Isolate Rhamnogalacturonan-II in Arabidopsis thaliana Seed Mucilage
Dayan Sanhueza and Susana Saez-Aguayo
Sep 5, 2025 1248 Views

Detailed Method for the Purification of Rhamnogalacturonan-I (RG-I) in Arabidopsis thaliana
Liang Zhang [...] Breeanna R. Urbanowicz
Feb 5, 2026 144 Views
Abstract
The plant cell wall is primarily composed of the polysaccharides cellulose, hemicellulose and pectin. The structural and compositional complexity of these components are important for determining cell wall function during plant growth. Moreover, cell wall structure defines a number of functional properties of plant-derived biomass, such as rheological properties of foods and feedstock suitability for the production of cellulosic biofuels. A typical characterization of cell wall chemistry in the molecular biology lab consists of a mild acid hydrolysis for the quantification of hemicellulose and pectin-derived monomers and a separate analysis of cellulose by the Updegraff method. We have adopted a streamlined ‘one-step two-step’ hydrolysis protocol that allows for the simultaneous determination of cellulose content, neutral sugars, and uronic acids by high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) of paired samples. In our work, this protocol has largely replaced Updegraff cellulose quantification and hydrolysis with 2 M TFA for the determination of matrix polysaccharide composition at the micro scale.
Keywords: CelluloseBackground
The protocol is based on paired analysis of samples hydrolyzed in 4% (w/v) sulfuric acid at 121 °C. One set of samples is first pretreated with 72% (w/w) sulfuric acid to swell cellulose and make it susceptible to dilute acid hydrolysis (Saeman hydrolysis in Figure 1; Saeman et al., 1945). The other set of samples are not subjected to this pretreatment, resulting in hydrolysis of non-crystalline matrix polysaccharides (Matrix hydrolysis in Figure 1). Comparison of the glucose recovered from each hydrolysis regime allows calculation of cellulose amount that is in good agreement with the more labor-intensive Updegraff (1969) protocol (Bauer and Ibáñez, 2014). In addition to glucose (Glc), other sugars derived from matrix polysaccharides can be quantified from the matrix hydrolysis samples (Gao et al., 2014). Thus, with relatively few manual manipulations, matrix monosaccharides and cellulose can be quantified from two hydrolysis samples and a total of four HPAEC-PAD experiments. Despite the number of chromatographic separations required by the protocol, the great reduction in ‘hands-on’ time required to prepare samples makes this technique well-suited to high throughput analyses. Although we have found that robust and reproducible analysis of rhamnose (Rha), arabinose (Ara), mannose (Man), and xylose (Xyl) requires multiple HPAEC-PAD runs, reports describing simultaneous quantification of all neutral and acidic sugars in a single run may allow further improvement of this protocol’s throughput (Zhang et al., 2012; Voiniciuc and Grünl, 2016). The steps of the protocol described here are outlined in Figure 1.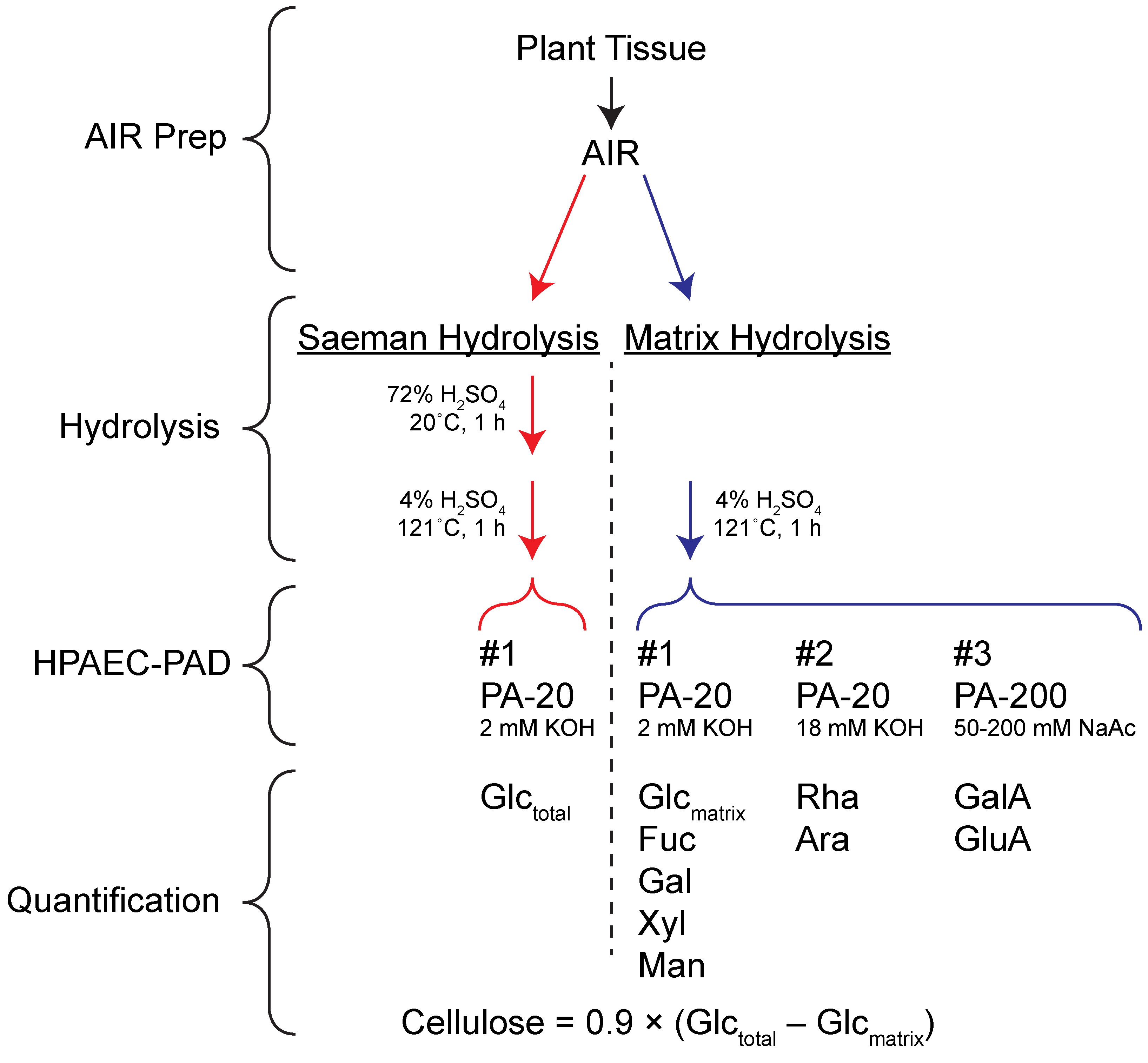
Figure 1. Protocol Overview. AIR = Alcohol insoluble residue.
Materials and Reagents
- 2 ml safe-lock microcentrifuge tubes (Eppendorf, catalog number: 022363352 )
- Aluminum foil
- 2 ml Sarstedt tubes with screw caps (SARSTEDT, catalog number: 72.694.007 )
- 2 ml screw cap autosampler vials (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: C4000-1W )*
- Autosampler vial caps with pre-slit septa (Phenomenex, catalog number: AR0-8977-13-B )*
- 5/32” Grinding Balls, 440C Stainless Steel, Treated (OPS Diagnostics, catalog number: GBSS 156-5000-01 )*
- Disposable anti-static polypropylene powder scoops (Cole-Parmer Instrument, catalog number: 06277-60 )
Note: This product has been discontinued. - Ethanol (Decon Labs, catalog number: V1016 )*
- Chloroform (Thermo Fisher Scientific, Fisher Scientific, catalog number: C607-4 )*
- Methanol (Thermo Fisher Scientific, Fisher Scientific, catalog number: A412P4 )*
- Acetone (Thermo Fisher Scientific, Fisher Scientific, catalog number: A184 )*
- 72% (w/w) sulfuric acid solution (RICCA Chemical, catalog number: R81916001A )*
- Ultrapure water (Milli-Q or equivalent)*
- 9 sugars:
L-fucose (Fuc) (Sigma-Aldrich, catalog number: F2252 )*
D-glucose (Glc) (Sigma-Aldrich, catalog number: G8270 )*
D-galactose (Gal) (Sigma-Aldrich, catalog number: G0750 )*
D-xylose (Xyl) (Sigma-Aldrich, catalog number: X1500 )*
D-mannose (Man) (Sigma-Aldrich, catalog number: M8574 )*
L-arabinose (Ara) (Sigma-Aldrich, catalog number: A3256 )*
L-rhamnose (Rha) (Sigma-Aldrich, catalog number: W373011 )*
D-galacturonic acid monohydrate (GalA) (Sigma-Aldrich, catalog number: 48280 )*
D-glucuronic acid (GluA) (Sigma-Aldrich, catalog number: G5269 )* - Sodium hydroxide solution (50%, w/w) (Thermo Fisher Scientific, Fisher Scientific, catalog number: SS254 )
- Sodium acetate, anhydrous (Sigma-Aldrich, catalog number: 71183 )
- Liquid nitrogen
*Note: These items can reliably be substituted with laboratory-grade equivalents from different vendors.
Equipment
- Freeze-dryer (Labconco, model: FreeZone 12 )* (optional)
- Magnet
- Aspirator
- Metal spatula
- Microcentrifuge (Eppendorf, model: 5417R )*
- Autoclave-compatible rack
- Autosampler
- Microbalance (Mettler Toledo, model: XS105 )*
- Ball mill (Retsch, model: MM 400 )*
- Dionex ICS-5000 HPAEC-PAD system, optionally equipped with an eluent generator (Thermo Fisher Scientific, Fisher Scientific, model: ICS-5000+ SYSTEM )
- CarboPac PA-20 Analytical column, 3 x 150 mm (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 060142 )
- CarboPac PA-20 Guard column, 3 x 30 mm (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 060144 )
- CarboPac PA-200 Analytical column, 3 x 250 mm (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 062896 )
- CarboPac PA-200 Guard column, 3 x 50 mm (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 062895 )
*Note: These items can reliably be substituted with laboratory-grade equivalents from different vendors.
Software
- Chromeleon 7 (Thermo Fisher Scientific)
- Microsoft Excel
Procedure
- Preparation of alcohol insoluble residue (AIR)
- Flash freeze < 300 mg fresh tissue in a 2 ml Eppendorf tube (Note 1). Homogenization and solvent extractions will be inefficient if more tissue is used. If plants are derived from agar-solidified plates, care must be taken to avoid taking any agar with the seedlings, as it will be hydrolyzed and quantified as Gal in later steps. If a freeze-dryer is not available, see Note 2 and skip to step A6.
- Leave the lid of the tube open, but cover the opening with a small piece of aluminum foil. Ensure the foil is vented by poking a small hole with a needle or pipet tip.
- Lyophilize frozen samples for 2-3 days.
- Add 3 steel balls to each tube and close the lids.
- Homogenize the dried samples for 2 min, shaking at 25 Hz at room temperature. Reverse the orientation of the tube holder and shake for an additional 2 min at 25 Hz.
- Ensure that a fine powder with no clumps is obtained before proceeding.
- Add 1.5 ml of 70% (v/v) ethanol to the tube and vortex. Remove the steel balls from the slurry using a magnet and discard them.
- Centrifuge at 20,000 x g for 10 min.
- Discard the supernatant using an aspirator that is compatible with organic solvents or pipet.
- Add 1.5 ml of 1:1 (v:v) chloroform:methanol. Disrupt the pellet with a small metal spatula and then thoroughly vortex the sample.
- Repeat steps A8-10 two more times for a total of 3 washes with chloroform:methanol.
- Centrifuge and remove the solvents as before (steps A8-9).
- Add 1.5 ml of acetone, disrupt the pellet, vortex, centrifuge, and discard the supernatant as before.
- Dry the final pellet either by using a vacuum centrifuge for at least 30 min or let air-dry overnight. The resulting material is the AIR (Note 3, Figure 2).

Figure 2. AIR material after step A14
- Flash freeze < 300 mg fresh tissue in a 2 ml Eppendorf tube (Note 1). Homogenization and solvent extractions will be inefficient if more tissue is used. If plants are derived from agar-solidified plates, care must be taken to avoid taking any agar with the seedlings, as it will be hydrolyzed and quantified as Gal in later steps. If a freeze-dryer is not available, see Note 2 and skip to step A6.
- One-step two-step hydrolysis
- For each AIR sample, accurately weigh ~1 mg of material into each of four 2 ml Sarstedt tubes on an analytical balance (Note 4). Avoid clumps of material. Record the exact weight of AIR in each tube. These four tubes are technical replicates that will be subjected to two different hydrolysis regimes as duplicates.
- Saeman hydrolysis samples
- To two tubes containing AIR, add 50 μl of 72% (w/w) sulfuric acid. Immediately cap the tubes, vortex vigorously, and centrifuge briefly to collect the sample in the bottom of the tube. Delayed or insufficient initial mixing can result in poor rehydration of the sample and incomplete hydrolysis.
- Incubate the tube at room temperature for 1 h. Vortex every 10 min or continuously with an Eppendorf Thermomixer.
- Add 1,400 μl water to each tube to adjust the sulfuric acid concentration to 4% (w/v). Vortex to mix.
- Matrix hydrolysis samples
- To the second set of two tubes, add 1,400 μl water first and then add 50 μl 72% (w/w) sulfuric acid to give a 4% (w/v) concentration of sulfuric acid. Cap the tubes and vortex.
- Recovery standards
- Prepare a standard mix consisting of 9 sugars (Fuc, Rha, Ara, Gal, Glc, Xyl, Man, GalA, and GluA), each at a concentration of 100 µg ml-1. It is recommended to prepare a concentrated stock of each sugar individually and then combine. Account for the extra mass of water if hydrated sugars are used. Prepare at least enough for use in this part of the protocol (1 ml) as well as for preparation of a standard curve in the next part of the protocol (138 μl). Sugar standards can be stored frozen once prepared.
- To account for sugar-specific losses during hydrolysis, a recovery standard is subjected to the same conditions as the matrix hydrolysis samples (Note 5). Pipet 500 μl of the standard mixture and 900 μl of water into a 2 ml Sarstedt tubes. Add 50 μl of 72% (w/w) sulfuric acid. Repeat with a second tube. Cap the tubes and vortex.
- Prepare a standard mix consisting of 9 sugars (Fuc, Rha, Ara, Gal, Glc, Xyl, Man, GalA, and GluA), each at a concentration of 100 µg ml-1. It is recommended to prepare a concentrated stock of each sugar individually and then combine. Account for the extra mass of water if hydrated sugars are used. Prepare at least enough for use in this part of the protocol (1 ml) as well as for preparation of a standard curve in the next part of the protocol (138 μl). Sugar standards can be stored frozen once prepared.
- Set all tubes except for one of the recovery standards in an autoclave-compatible rack and autoclave at 121 °C for 60 min. The second recovery standard tube is not autoclaved and serves as a control for calculating monosaccharide-specific correction factors.
- Cool samples to room temperature and centrifuge for 1 min at 20,000 x g to pellet any insoluble material.
- Dilute samples, recovery standard, and recovery standard control 100 fold by transferring 10 μl into an autosampler vial containing 990 μl of MilliQ water. Store samples at 4 °C for up to two weeks. Samples can be stored for a month or more if they are frozen at -20 °C. This dilution factor is convenient for AIR isolated from Arabidopsis seedlings, for more cellulose-rich AIR prepared from stems, wood, or other secondary tissues, a higher dilution factor should be used (200-500 fold).
- Standard curve samples
- Dilute standard mix from step B4 (9 sugars, 100 µg ml-1 each) to yield a standard curve between 0.05 µg ml-1 and 5 µg ml-1. Prepare this directly into 2 ml autosampler vials with pre-slit septa (Table 1). This range of concentrations generally covers the complete linear range of the instrument.
Table 1. Preparation of standard curve samples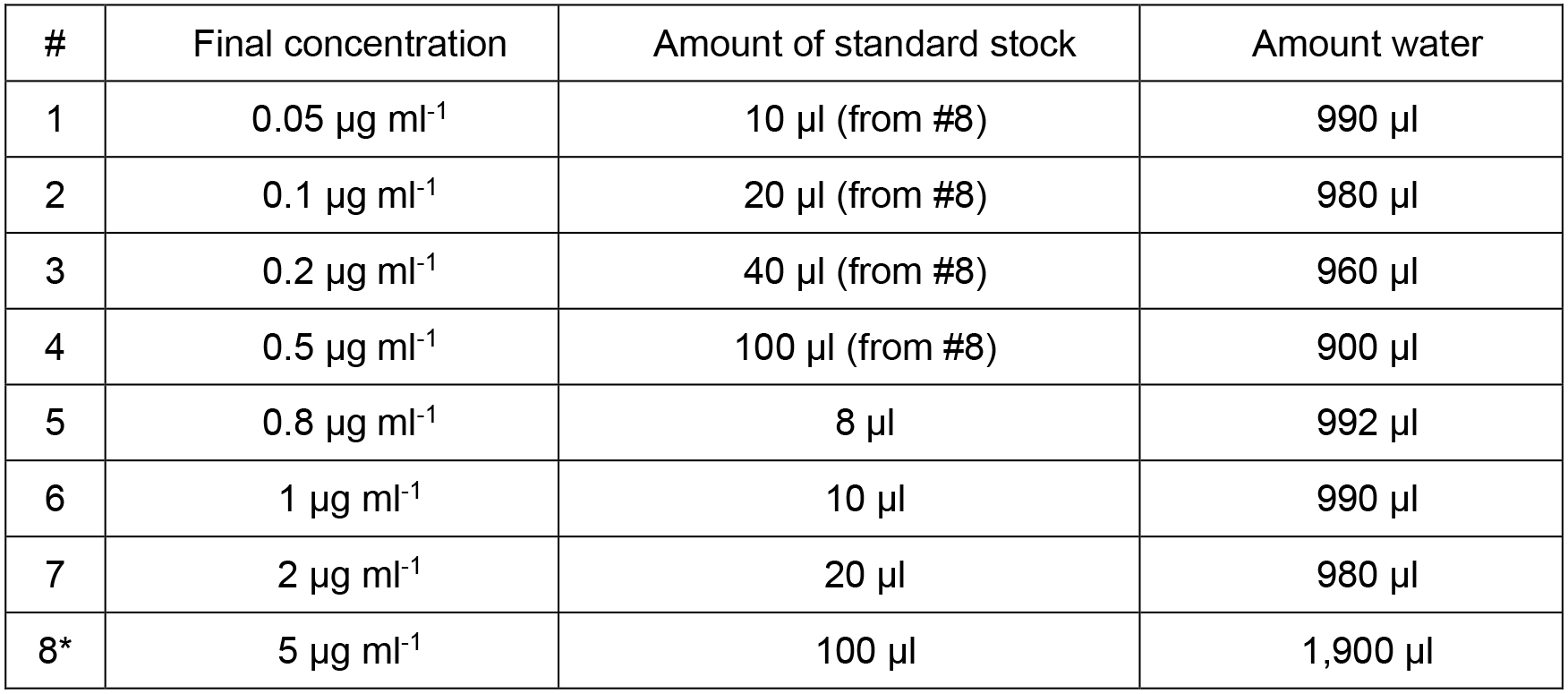
*Note: Standard 8 is further diluted to prepare standards 1-4.
- Dilute standard mix from step B4 (9 sugars, 100 µg ml-1 each) to yield a standard curve between 0.05 µg ml-1 and 5 µg ml-1. Prepare this directly into 2 ml autosampler vials with pre-slit septa (Table 1). This range of concentrations generally covers the complete linear range of the instrument.
- For each AIR sample, accurately weigh ~1 mg of material into each of four 2 ml Sarstedt tubes on an analytical balance (Note 4). Avoid clumps of material. Record the exact weight of AIR in each tube. These four tubes are technical replicates that will be subjected to two different hydrolysis regimes as duplicates.
- HPAEC-PAD analysis
- HPAEC-PAD analysis #1 (All samples)
- Inject 25 μl of each standard, recovery standard, and sample onto a 3 x 150 mm CarboPac PA-20 analytical column equipped with a 3 x 50 mm guard column of the same material. Elute the compounds with 2 mM KOH at a flow rate of 0.4 ml min-1 for 30 min (Note 6). This run will resolve Fuc, Gal, Glc, Xyl, and Man. Rha and Ara are not consistently separated with these conditions, so they are quantified by a second analysis that is only necessary for the matrix hydrolysis samples.
- In our experience, retention times of peaks will shift earlier as more samples are run, and after ~50-100 samples it is necessary to perform a column flush with sodium acetate (see below, step C4).
- Inject 25 μl of each standard, recovery standard, and sample onto a 3 x 150 mm CarboPac PA-20 analytical column equipped with a 3 x 50 mm guard column of the same material. Elute the compounds with 2 mM KOH at a flow rate of 0.4 ml min-1 for 30 min (Note 6). This run will resolve Fuc, Gal, Glc, Xyl, and Man. Rha and Ara are not consistently separated with these conditions, so they are quantified by a second analysis that is only necessary for the matrix hydrolysis samples.
- HPAEC-PAD analysis #2 (Matrix hydrolysis samples only)
- Inject 25 μl of each standard, recovery standard, and sample onto a 3 x 150 mm CarboPac PA-20 analytical column equipped with a 3 x 50 mm guard column of the same material. Elute the compounds with 18 mM KOH 0.4 ml min-1 for 30 min. This run will resolve Rha and Ara.
- A column flush may be required after ~50-100 samples are run (see below, step C4).
- Inject 25 μl of each standard, recovery standard, and sample onto a 3 x 150 mm CarboPac PA-20 analytical column equipped with a 3 x 50 mm guard column of the same material. Elute the compounds with 18 mM KOH 0.4 ml min-1 for 30 min. This run will resolve Rha and Ara.
- HPAEC-PAD analysis #3 (Matrix hydrolysis samples only)
- Inject 25 μl of each standard, recovery standard, and sample onto a 3 x 150 mm CarboPac PA-200 analytical column equipped with a 3 x 50 mm guard column of the same material. Elute the compounds with a 10 min gradient of 50 mM to 200 mM sodium acetate in 100 mM NaOH at a flow rate of 0.4 ml min-1 (This is accomplished with a gradient of 5%-20% eluent B [100 mM NaOH, 1 M sodium acetate]. Eluent A consists of 100 mM NaOH and makes up the remaining volume). This run will resolve GalA and GluA.
- Inject 25 μl of each standard, recovery standard, and sample onto a 3 x 150 mm CarboPac PA-200 analytical column equipped with a 3 x 50 mm guard column of the same material. Elute the compounds with a 10 min gradient of 50 mM to 200 mM sodium acetate in 100 mM NaOH at a flow rate of 0.4 ml min-1 (This is accomplished with a gradient of 5%-20% eluent B [100 mM NaOH, 1 M sodium acetate]. Eluent A consists of 100 mM NaOH and makes up the remaining volume). This run will resolve GalA and GluA.
- Column flushing procedure
- The performance of the CarboPac PA20 column degrades after running ~50-100 samples, leading to increasingly early retention times until peaks cannot be sufficiently resolved. This is remedied by flushing the column with 100 mM NaOH/1 M sodium acetate for 30 min followed by water for 30 min at 0.2 ml min-1. If the CarboPac PA20 column is being used with an HPAEC-PAD system that is equipped with an eluent generator, it will need to be temporarily moved to a different system or port in order to complete this procedure.
- HPAEC-PAD analysis #1 (All samples)
Data analysis
- Integrate all standard curve and hydrolysis samples using Chromeleon 7 software. For the calibration curves, a quadratic curve that is forced through the origin is used. For some samples, peak identity may need to be manually assigned as retention times tend to shift earlier with multiple runs. Figures 3-5 show representative chromatograms from the three HPAEC-PAD methods.
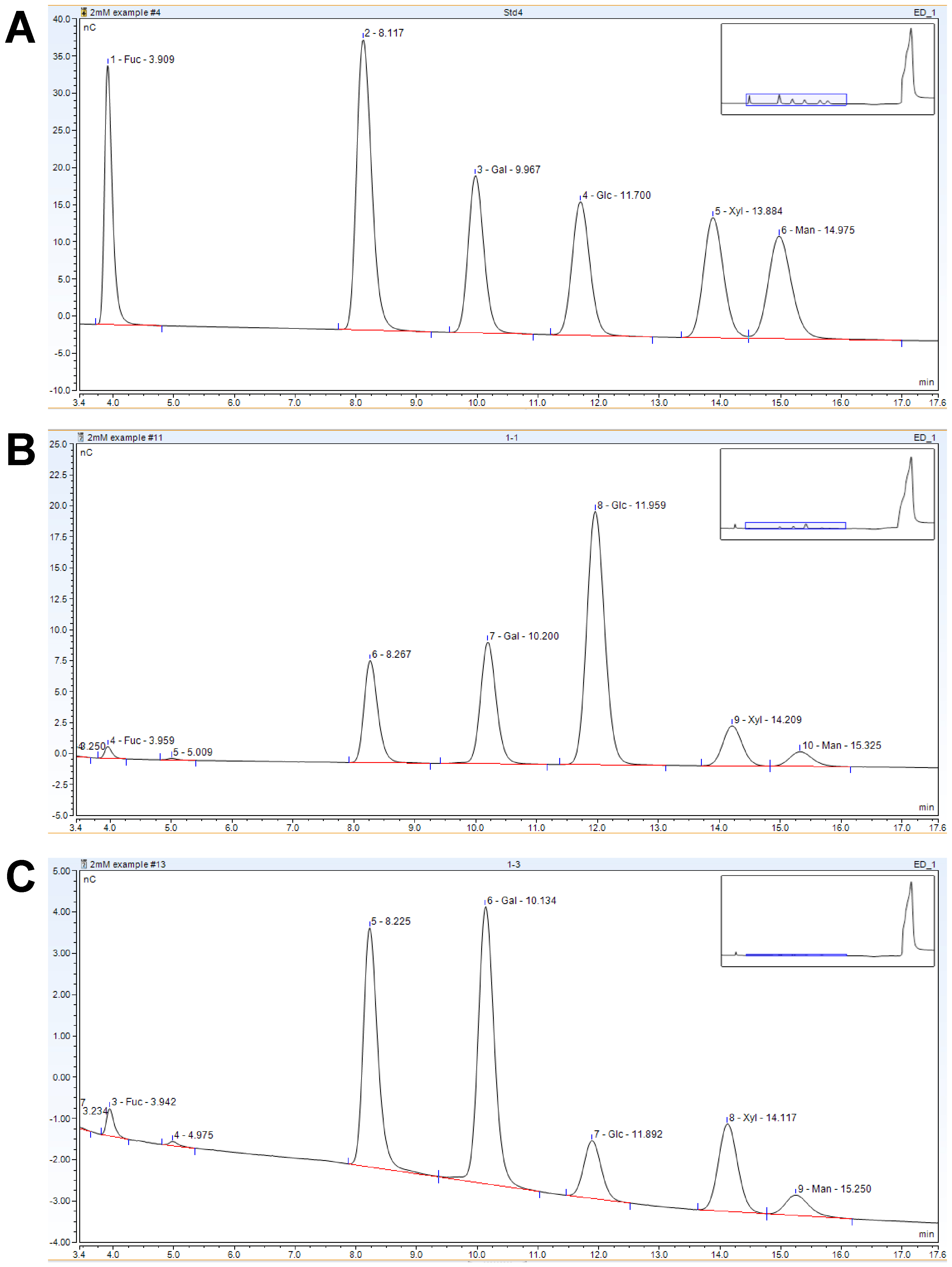
Figure 3. HPAEC-PAD analysis #1 representative chromatograms. A. Standard mixture. B. Saeman hydrolysis sample. C. Matrix hydrolysis sample. The peak at ~8.2 min contains both Rha and Ara and is not quantified in this method.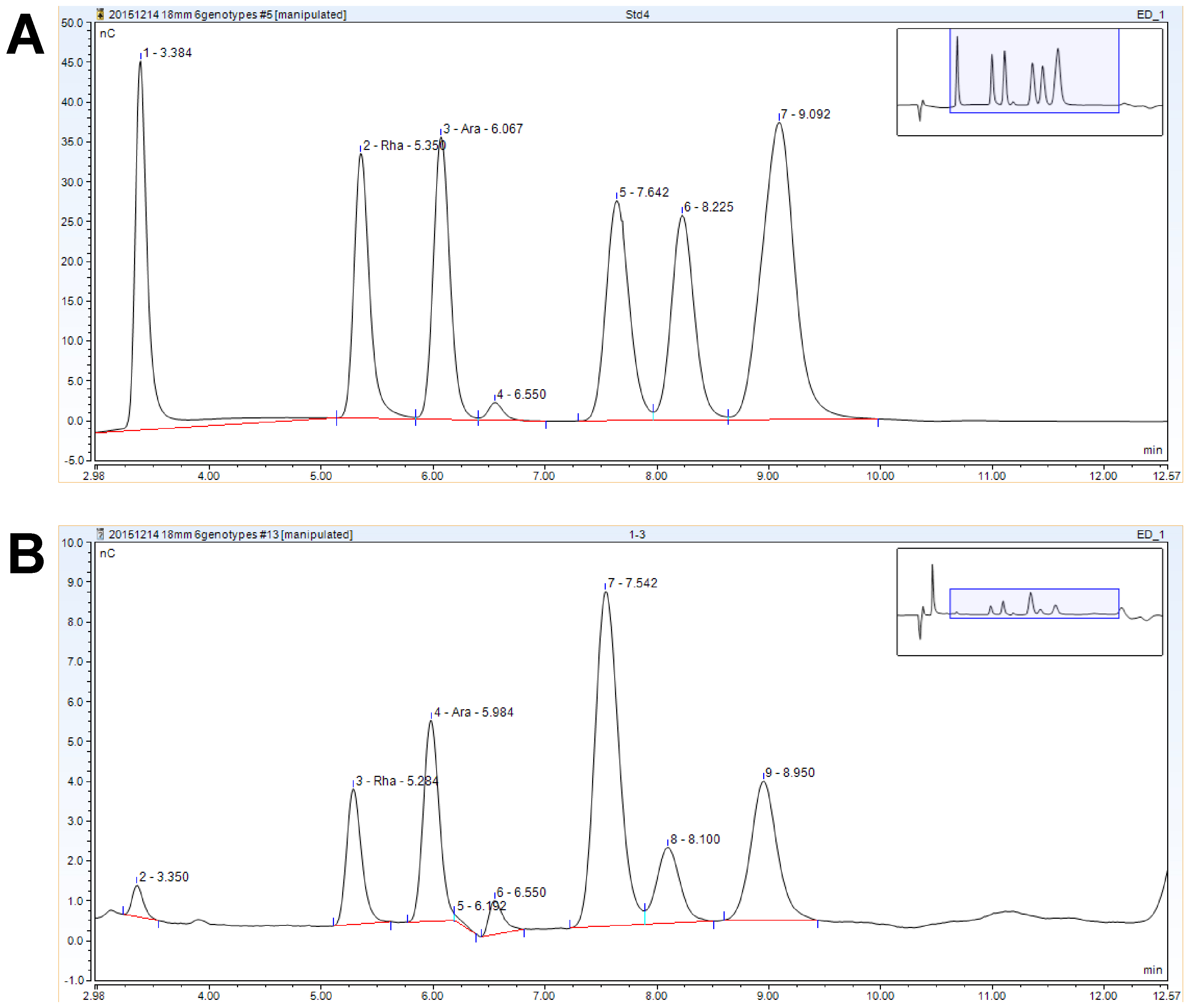
Figure 4. HPAEC-PAD analysis #2 representative chromatograms. A. Standard mixture. B. Matrix hydrolysis sample. Only Rha and Ara are quantified in these runs. Fuc elutes at ~3.4 min, Gal elutes at ~7.6 min, Glc elutes at ~8.2 min. The peak at ~9.0 min is a mixture of Xyl and Man.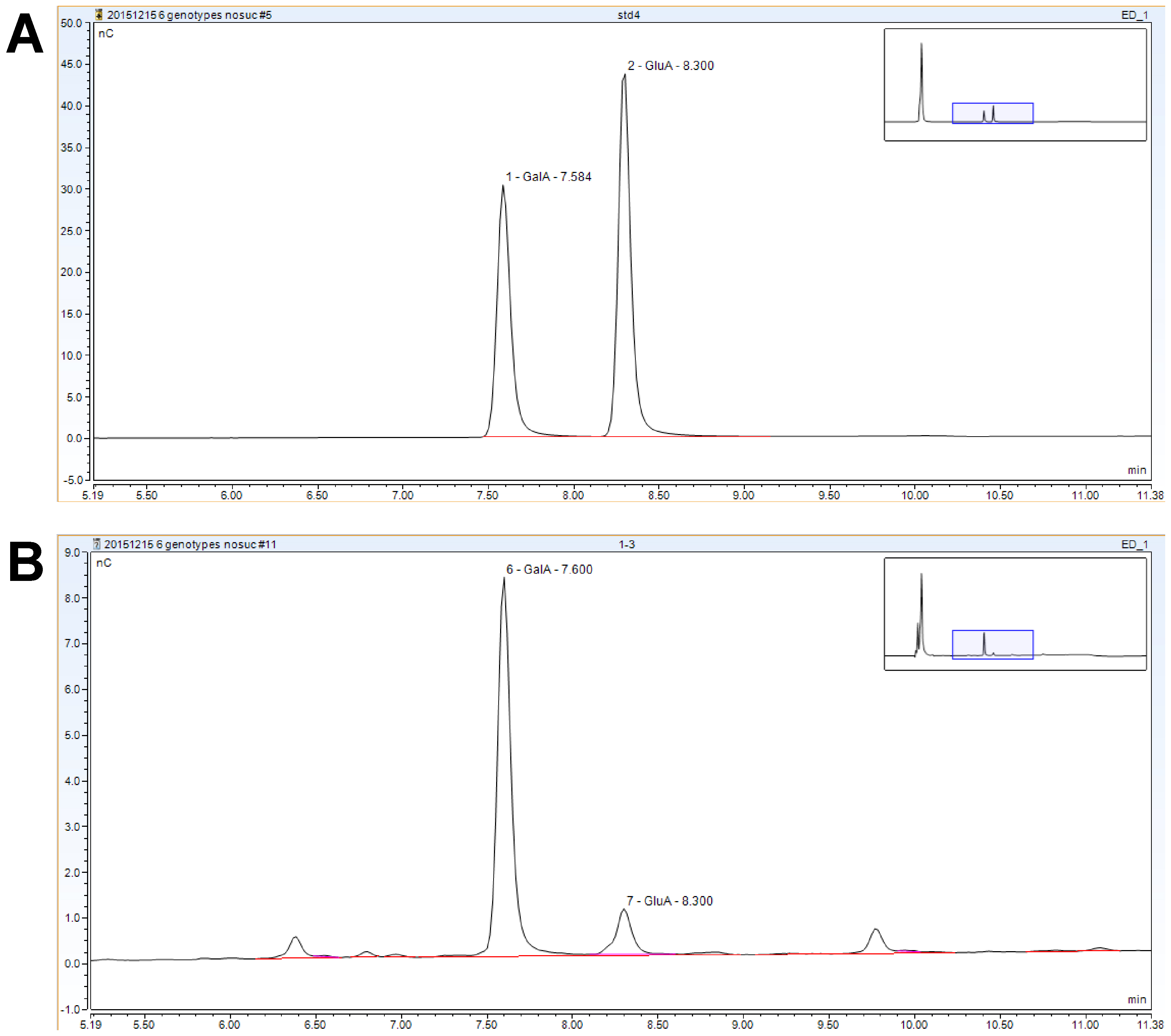
Figure 5. HPAEC-PAD analysis #3 representative chromatograms. A. Standard mixture. B. Matrix hydrolysis sample. - From each run, the results (assayed monosaccharide concentration, µg ml-1) can be copied from the results table into a Microsoft Excel spreadsheet. Set up a table with results for each sugar in columns and a column with the AIR mass for each sample (Figure 6).
- Calculate correction factors for each sugar by dividing the concentration of the recovery standard by the concentration of the respective sugar in the recovery standard control. These are typically > 0.85 for neutral sugars, but are lower for uronic acids (~0.6-0.8). The same Glc recovery factor is used for both Saeman and Matrix hydrolysis samples. Lower than expected correction factors indicate excessive sample loss that can occur due to prolonged high temperature treatment. Repeat the hydrolysis and ensure that samples are cooled and diluted as soon as the autoclave run is complete.
- In a new column, calculate the recovery-standard corrected monosaccharide amounts as µg per mg of AIR using the following formula:

Where,
c = Assayed concentration (µg ml-1),
d = 100 (dilution factor),
v = 1.450 (volume, ml),
r = Recovery correction factor (From step C3),
m = AIR mass (mg). - At this point, technical replicates can be validated to check if there is any systematic difference in monosaccharide quantification that would justify discarding a technical replicate. (This would be indicative of sample loss or a weighing error.). Fuc, Gal, and Xyl should have consistent values in both the Saeman and Matrix hydrolysis samples. Glc will be much higher in the Saeman hydrolysis samples, and Man is expected to be more abundant in these samples as well since Glc isomerization to Man is a known side reaction during Saeman hydrolysis (Carpita and Shea, 1989). The extent of this side reaction is only ~2%, so it is ignored for the purpose of cellulose quantification.
- Average technical replicates into a new column. Only the Matrix hydrolysis samples, which were subjected to all three HPAEC-PAD methods, are used for reporting neutral sugars and uronic acids.
- To calculate cellulose amount, subtract Glcmatrix (from Matrix hydrolysis samples) from Glctotal (from Saeman hydrolysis samples) and multiply by 0.9. The 0.9 correction factor reflects the difference in molar mass between glucose (180.16 g mol-1) and anhydroglucose (162.14 g mol-1), as it occurs in the cellulose polymer prior to hydrolysis.
Cellulose (µg mg-1 AIR) = 0.9 × (Glctotal (µg mg-1) - Glcmatrix (µg mg-1)) - For statistical analysis, we typically use at least three biological replicates of AIR prepared from independent plants or pools of seedlings. Results are reported as means and standard deviation and statistical significance is assessed by Student’s t-test or one-way ANOVA followed by Tukey’s multiple comparison test.
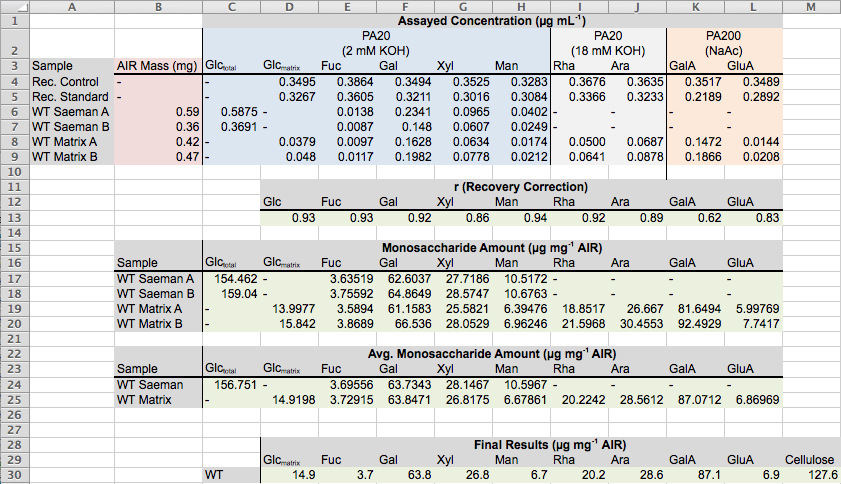
Figure 6. Screenshot of Excel spreadsheet illustrating the calculation of monosaccharide and cellulose from HPAEC-PAD integration data. The sample analyzed was AIR prepared from 5d old dark-grown Arabidopsis seedlings.
Notes
- Any starch present in the sample will contribute to glucose quantified in both hydrolysis regimes. If quantification of glucose derived from matrix polysaccharides is desired, starch can be reduced by moving light-grown plants to the dark for 24 h before tissue is collected (Sorek et al., 2015). Alternatively, starch can be enzymatically removed during preparation of the AIR (Pettolino et al., 2012) or can be independently assayed from the same AIR material (Yeats et al., 2016). Starch will not impact the determination of cellulose since it will contribute equally to the glucose quantified in the Saeman and matrix hydrolysis samples.
- If a freeze-dryer is not available, it is possible to prepare AIR directly from frozen tissue that is homogenized with a mortar and pestle or a ball mill. In both cases, the sample must remain frozen during homogenization: the mortar and pestle or ball mill tube rack should be immersed in liquid nitrogen before use. Tubes are more brittle at low temperatures and we have noted an increased rate of tubes breaking during homogenization in the ball mill, so we prefer to work with dried tissue at room temperature.
- The terms AIR and cell wall material are often used interchangeably, however, it is clear from the mass balance of quantified monosaccharides that AIR prepared from seedlings is only ~50% cell wall material. More extensive washing of AIR with an aqueous solution of SDS and sodium metabisulfite removes additional non-cell wall material (Dick-Pérez et al., 2011). However, this protocol is lengthy and inconvenient if multiple samples are to be analyzed. Since there can be variation in the efficiency of extraction and washing during AIR preparation, it is best practice to prepare all AIR samples to be analyzed at the same time. We find it is convenient to prepare up to ~60 AIR samples simultaneously, with centrifuge capacity being a limiting factor. For hydrolysis steps, the number of tubes multiplies by 4, so once AIR is prepared, we proceed in batches of 15-20 samples (a total of 60-80 hydrolysis samples). For a large project, we recommend including a control sample with each batch.
- Handling the dry AIR material can be difficult due to the buildup of static charge. For moving the AIR to Sarstedt tubes, we use disposable anti-static polypropylene powder scoops (Cole-Parmer). Laboratory humidity > 40% will help to reduce static.
- Sugar losses occurring during hydrolysis are somewhat concentration dependent. For convenience, a standard mixture with equal amounts of each sugar is prepared. For more accurate absolute quantification, recovery standards can be prepared based on the expected composition of the samples being analyzed.
- LC elution with the same concentration of NaOH instead of KOH gives comparable results. In our work, we have used an HPAEC-PAD equipped with a KOH eluent generator module for the CarboPac PA-20 analyses. User-prepared eluents are typically prepared with NaOH. The procedure for eluent preparation is detailed in the CarboPac PA-20 manual. Following the outlined procedure is particularly important for avoiding carbonate entering the system, which drastically affects the chromatography of sugars.
Acknowledgments
We thank Will Barnes (Penn State University) and Chris Somerville (University of California, Berkeley) for helpful comments on this protocol. Funding for this work is from the Energy Biosciences Institute and the Philomathia Foundation. This protocol is based on the following previously published reports: Bauer and Ibáñez, 2014; Chen et al., 2016; Sorek et al., 2015; and Yeats et al., 2016.
References
- Bauer, S. and Ibanez, A. B. (2014). Rapid determination of cellulose. Biotechnol Bioeng 111(11): 2355-2357.
- Carpita, N. C. and Shea, E. M. (1988). Linkage structure of carbohydrates by gas chromatography-mass spectrometry (GC-MS) of parially methylated alditol acetates. In Biermann, C. J. and McGinnis, G. D. (Eds). Analysis of carbohydrates by GLC and MS. CRC Press, 157-216.
- Chen, S., Jia, H., Zhao, H., Liu, D., Liu, Y., Liu, B., Bauer, S. and Somerville, C. R. (2016). Anisotropic cell expansion is affected through the bidirectional mobility of cellulose synthase complexes and phosphorylation at two critical residues on CESA3. Plant Physiol 171(1): 242-250.
- Dick-Perez, M., Zhang, Y., Hayes, J., Salazar, A., Zabotina, O. A. and Hong, M. (2011). Structure and interactions of plant cell-wall polysaccharides by two- and three-dimensional magic-angle-spinning solid-state NMR. Biochemistry 50(6): 989-1000.
- Gao, X., Kumar, R. and Wyman, C. E. (2014). Fast hemicellulose quantification via a simple one-step acid hydrolysis. Biotechnol Bioeng 111(6): 1088-1096.
- Pettolino, F. A., Walsh, C., Fincher, G. B. and Bacic, A. (2012). Determining the polysaccharide composition of plant cell walls. Nat Protoc 7(9): 1590-1607.
- Saeman, J. F., Bubl, J. L. and Harris, E. E. (1945). Quantitative saccharification of wood and cellulose. Ind Eng Chem Anal Ed 17: 35-37.
- Sorek, N., Szemenyei, H., Sorek, H., Landers, A., Knight, H., Bauer, S., Wemmer, D. E. and Somerville, C. R. (2015). Identification of MEDIATOR16 as the Arabidopsis COBRA suppressor MONGOOSE1. Proc Natl Acad Sci U S A 112(52): 16048-16053.
- Updegraff, D. M. (1969). Semimicro determination of cellulose in biological materials. Anal Biochem 32(3): 420-424.
- Voiniciuc, C. and Günl, M. (2016). Analysis of monosaccharides in total mucilage extractable from Arabidopsis seeds. Bio-protocol 6(9): e1801.
- Yeats, T. H., Sorek, H., Wemmer, D. E. and Somerville, C. R. (2016). Cellulose deficiency is enhanced on hyper accumulation of sucrose by a H+-coupled sucrose symporter. Plant Physiol 171(1): 110-124.
- Zhang, Z., Khan, N. M., Nunez, K. M., Chess, E. K. and Szabo, C. M. (2012). Complete monosaccharide analysis by high-performance anion-exchange chromatography with pulsed amperometric detection. Anal Chem 84(9): 4104-4110.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Yeats, T., Vellosillo, T., Sorek, N., Ibáñez, A. B. and Bauer, S. (2016). Rapid Determination of Cellulose, Neutral Sugars, and Uronic Acids from Plant Cell Walls by One-step Two-step Hydrolysis and HPAEC-PAD. Bio-protocol 6(20): e1978. DOI: 10.21769/BioProtoc.1978.
- Yeats, T. H., Sorek, H., Wemmer, D. E. and Somerville, C. R. (2016). Cellulose deficiency is enhanced on hyper accumulation of sucrose by a H+-coupled sucrose symporter. Plant Physiol 171(1): 110-124.
Category
Plant Science > Plant biochemistry > Carbohydrate
Biochemistry > Carbohydrate > Cellulose
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









