- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Imaging Thick Lymph Node Tissue Sections
Published: Vol 6, Iss 18, Sep 20, 2016 DOI: 10.21769/BioProtoc.1938 Views: 13625
Reviewed by: Ivan ZanoniFrancesca MingozziMeenal SinhaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Calcium Imaging in T Lymphocytes: a Protocol for Use with Genetically Encoded or Chemical Ca2+ Indicators
Amit Jairaman and Michael D. Cahalan
Oct 5, 2021 5016 Views

Three-dimensional Co-culture Model for Live Imaging of Pancreatic Islets, Immune Cells, and Neurons in Agarose Gel
Elke M. Muntjewerff [...] Gustaf Christoffersson
Oct 20, 2023 2623 Views

Measuring Piezo1 and Actin Polarity in Chemokine-Stimulated Jurkat Cells During Live-Cell Imaging
Chinky Shiu Chen Liu [...] Dipyaman Ganguly
Oct 5, 2024 1802 Views
Abstract
Our protocol describes a simple procedure for imaging thick lymph node sections by 2-photon microscopy. Lymph nodes are sectioned using a vibratome (vibrating microtome) to produce slices of tissue that can then be stained with fluorescently labeled antibodies. The thick tissue sections (150-200 μm depth) allow for the detection of cell clustering that is typically under-represented in thin sections (10-20 μm) used for conventional confocal microscopy. Application of 2-photon microscopy facilitates imaging through the thick volume of the vibratome sections. In combination with automated image processing software, a thick lymph node cross-section image also facilitates quantitation of cellular events within a relatively large area of the tissue, thus providing a clearer picture on the spatial distribution of cellular events of interest (e.g., T cell clustering). This method can also readily be applied to other tissues, such as the spleen or skin.
Keywords: 2-photon microscopyMaterials and Reagents
- Razor blades, single edge, No.9 (0.22 mm) (VWR International, catalog number: 55411-050 )
- Greiner CELLSTAR® 24-well flat-bottom plates (Sigma-Aldrich, catalog number: M8812 )
- 96-well round-bottom plates (Corning, catalog number: 3788 )
- Durapore tape (3M, catalog number: 1538-1 )
- SuperfrostTM Plus and ColorFrostTM glass slides (25 x 75 x 1.0 mm) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 4951PLUS4 )
- Coverslips, No.1.5 (ProSciTech, catalog number: G425-2460 )
- Agarose (Sigma-Aldrich, catalog number: A6013-250G )
- Vetbond tissue adhesive (3M, catalog number: 1469SB )
- Phosphate-buffered saline (PBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10010023 )
- Protein block (Dako, catalog number: X0909 )
- Normal donkey serum (NDS) (Jackson ImmunoResearch, catalog number: 017-000-121 )
- Prolong gold antifade reagent (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: P36930 )
- Antibodies [e.g., pacific blue anti-B220, RA3-6B2 (Biolegend, catalog number: 103230 ); goat anti-CD69 (R&D Systems, catalog number: AF2386 )]
- Sodium periodate (NaIO4) (Sigma-Aldrich, catalog number: 311448 )
- Sodium dihydrogen orthophosphate, monohydrate (NaH2PO4·H2O) (monobasic) (VWR, catalog number: 97062-412 )
- Di-sodium hydrogen orthophosphate, anhydrous (Na2HPO4) (dibasic) (Ajax Finechem, catalog number: 621-500G )
- L-lysine (Sigma-Aldrich, catalog number: L5501 )
- Gelatin from porcine skin (Sigma-Aldrich, catalog number: G1890-100G )
- Glycerol for molecular biology (≥ 99%) (Sigma-Aldrich, catalog number: G5516 )
- 16% paraformaldehyde aqueous solution (Electron Microscopy Sciences, catalog number: 30525-89-4 )
- Periodate-lysine-paraformaldehyde (PLP) fixative (see Recipes)
- P-buffer (see Recipes)
- L-lysine (see Recipes)
- 5% gelatin/glycerol mix (see Recipes)
Equipment
- Vibratome (Leica, model: VT1200S )
- Dumont No.5 forceps (Fine Science Tools, catalog number: 11251-10 )
- Scissors, straight, sharp (Roboz, catalog number: RS-6752 )
- Soft bristled paint brush (e.g., size 2/0)
- Microscope with tunable Coherent Chameleon Ti:Sa laser (ZEISS, model: LSM 710 NLO )
Note: This product has been discontinued (Replaceable item, e.g., ZEISS, model: LSM 800 ). - Scale for weighing 1 g
- Microwave
Software
- Imaris, Bitplane (http://www.bitplane.com/)
Procedure
- Harvest lymph nodes (LNs) and place in 2 ml of PLP or 2% paraformaldehyde (PFA) at 4 °C for fixation.
Notes:- This step is optional but crucial for preserving cellular morphology, but may compromise certain antibody staining. One example we have tested is anti-XCR1 (clone: ZET) where fixation abrogates staining. PLP is preferable for preserving fine cellular morphology, such as cell dendrites.
- All procedures involving animals must be performed according to approved institutional, regional or national protocols (as applicable) by appropriately trained and certified personnel.
- This step is optional but crucial for preserving cellular morphology, but may compromise certain antibody staining. One example we have tested is anti-XCR1 (clone: ZET) where fixation abrogates staining. PLP is preferable for preserving fine cellular morphology, such as cell dendrites.
- After fixation (4-6 h for small LNs, and up to 12 h for larger tissues, such as swollen LN after inflammation), transfer tissues into 2 ml of cold PBS and keep on ice.
- Embed tissues into agarose gel:
- Pour warm 2% agarose in PBS (boiled then cooled under running tap water until warm to touch) into wells in 24-well plates;
- Embed tissues into wells loaded with warm agarose and in desired orientation, Figure 1 (Each LN should touch the bottom of the well and will later be sliced parallel to this plane);
- Make sure the plate is level and leave on ice to allow agarose to set.

Figure 1. Embedding LN in agarose gel. Pour warm agarose into wells, embed LN into agarose and adjust to desired orientation (left). Allow agarose to set (right).
- Pour warm 2% agarose in PBS (boiled then cooled under running tap water until warm to touch) into wells in 24-well plates;
- Meanwhile, start up the vibratome and calibrate the cutting blade as per manufacturer’s instructions. Prepare an ice bath and fill the buffer tray with cold PBS, Figure 2.

Figure 2. Setting up vibratome. Insert blade and calibrate, prepare ice bath, and fill buffer tray with cold saline buffer. - Remove the agarose block containing the LN with a pair of forceps and carefully trim into cuboid shape with a razor blade, Figure 3.
- Apply 1-2 strips of 3M Durapore tape to the specimen holder. Attach the trimmed LN/agarose block on the specimen holder (with the LN near the top of the block) using a drop of Vetbond adhesive (Figure 3, right).
Note: Cyanoacrylate glue as recommended by the manufacturer is preferred. However, if Vetbond is used, wait ~1 min after tissue block is attached for the glue to dry.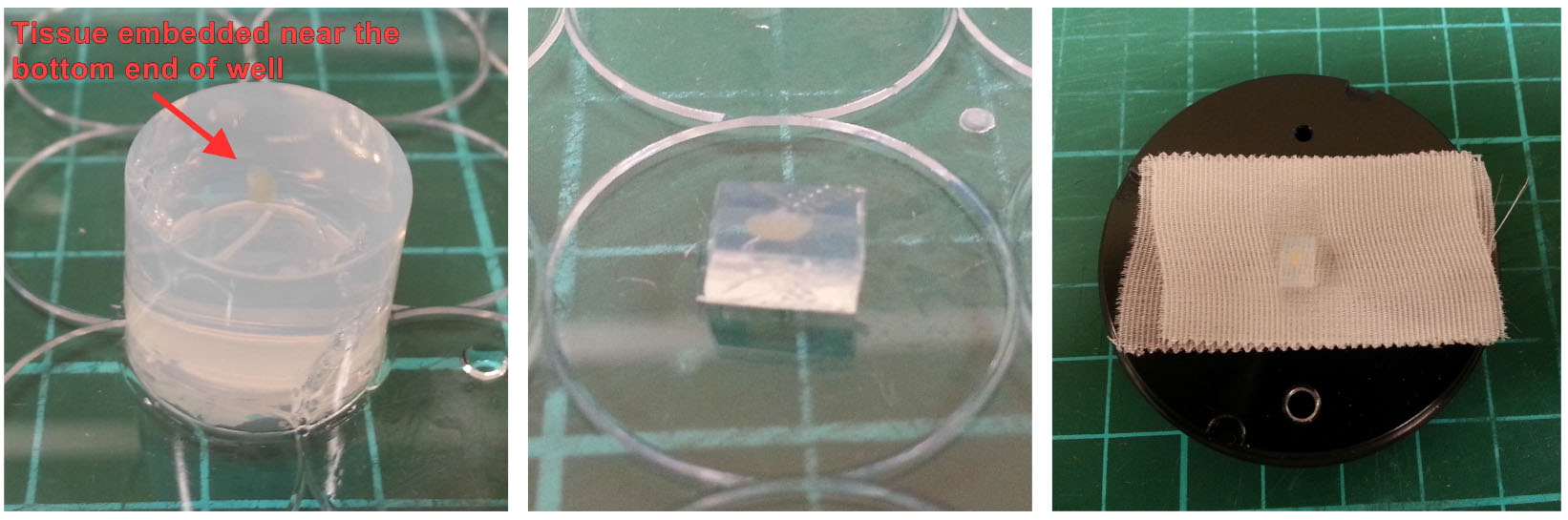
Figure 3. Mounting LN/agarose block. Remove set agarose gel from well (left), trim with a razor blade into cuboid shape (middle) and mount the block on to the specimen holder with a small amount of glue (right). - Mount the specimen holder onto vibratome for cutting. The LN should be aligned such that it is sectioned longitudinally as we find that tissue quality is better preserved (Figure 4).
- Set up the vibratome to ensure appropriate cutting speed and thickness. Typically, we use ~0.03 mm/s blade speed with ~1.6 mm amplitude, and ~200-250 μm thickness for best preservation of LN structural integrity.
- Begin with the vibratome blade just above the agarose surface, using brush as guide, and begin cutting via automated single or continuous sectioning. Discard the first slice.
- Transfer cut slices (with a No.5 forcep and soft bristled brush) into wells in a 96-well round-bottom plate filled with cold PBS, Figure 4. Take particular care to grip only the agarose part of the slice so as not to disrupt the LN tissue.
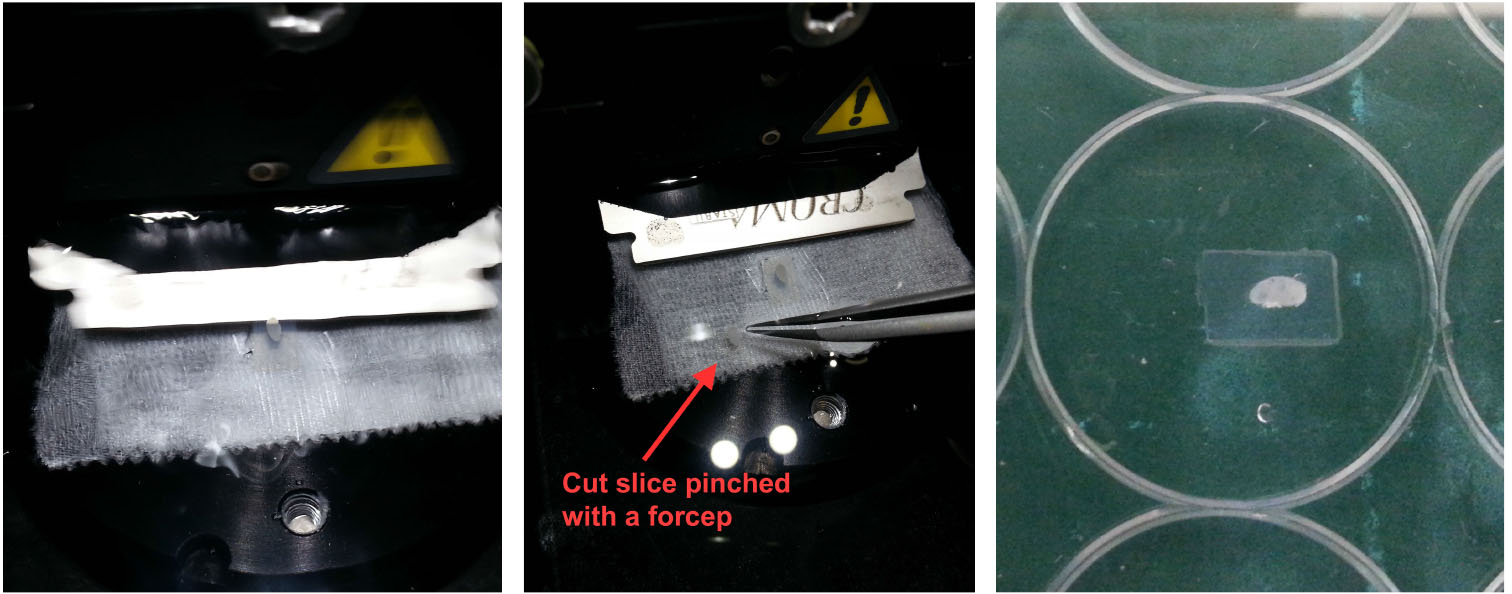
Figure 4. Handling vibratome slices. Once an LN/agarose slice has been cut (left), use a pair of fine forceps and a brush to retrieve the slice from the buffer tray (middle) and leave it in cold PBS. For reference, the slice should appear relatively intact with minimal disruption to the tissue (right). - Prepare a 96-well round-bottom plate for antibody staining by filling each row with reagents to be used in sequential steps for antibody staining (Figure 5):
Row 1: Cold PBS (where LN slices are initially placed)
Row 2: Protein block
Row 3: Primary antibodies (~70 μl per well)
Row 4: PBS (Wash #1)
Row 5: PBS (Wash #2)
Row 6: Secondary antibodies (~70 μl per well)
Row 7: PBS (Wash #1)
Row 8: PBS (Wash #2) - Transfer the LN slices down each column after each incubation step.
- Incubate in protein block for 1-2 h.
- Transfer into the well containing the primary antibodies (diluted in 2.5% NDS) and incubate at 4 °C for 8 h or overnight.
Note: We have tested Pacific Blue-conjugated anti-B220 (dilution 1:200) and unconjugated anti-CD69 (dilution 1:50). It should be noted that each antibody batch should be individually titrated for optimal results. - Transfer to the first wash buffer (PBS) well for 20-30 min, followed by the second well containing wash buffer (PBS).
- Transfer into the well containing the secondary antibodies (diluted in 2.5% NDS) and incubate at 4 °C for 8 h or overnight.
- Transfer to the first wash buffer (PBS) well for 20 min, followed by the second well containing wash buffer (PBS).
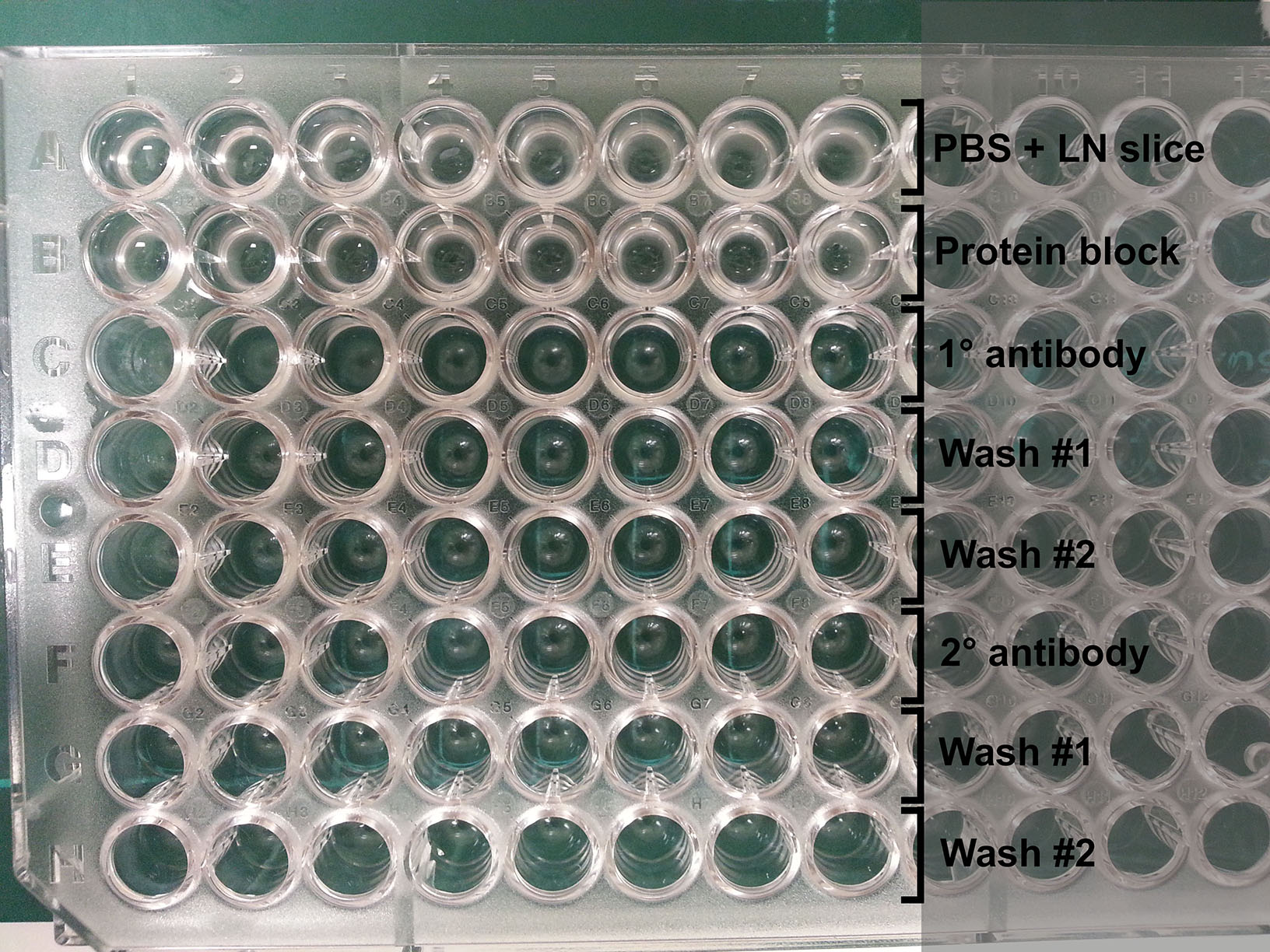
Figure 5. Plate setup for antibody staining. Each well contains a LN slice that is transferred down each column for antibody labeling and washing steps. - Place LN slices on glass microscope slides (up to 4 LN slices per slide). Arrange them in desired orientation.
- Use 5% gelatin/glycerol in PBS as mounting media and carefully place a coverslip over the tissues such that the edges are sealed.
- Slides are now ready for two-photon imaging:
Acquisition parameters differ and are dependent upon the desired quality and fluorophore configuration (see Notes).
In our experiments, we use: Zeiss LSM710 NLO with tunable Coherent Chameleon Ti:Sa laser Zeiss 20x/1.0 NA water immersion lens.
Resolution: 512 x 512
Slice interval: 2.5 μm
Zoom: ~1.5x
Depth-adjusted correction for laser power and gain settings to reduce excessive laser excitation of the tissue, which can lead to photobleaching and phototoxicity.
Whole LN cross-sections are acquired using the tile-scanning function of the Zeiss Zen software. An example is shown in Figure 6. - Post-acquisition processing can be performed on dedicated image processing software (e.g., Bitplane Imaris) to allow quantitation of cells (Spots function) and export of statistics. Additionally, Imaris XTension functions or customized MATLAB script can be used for further analysis.
Representative data
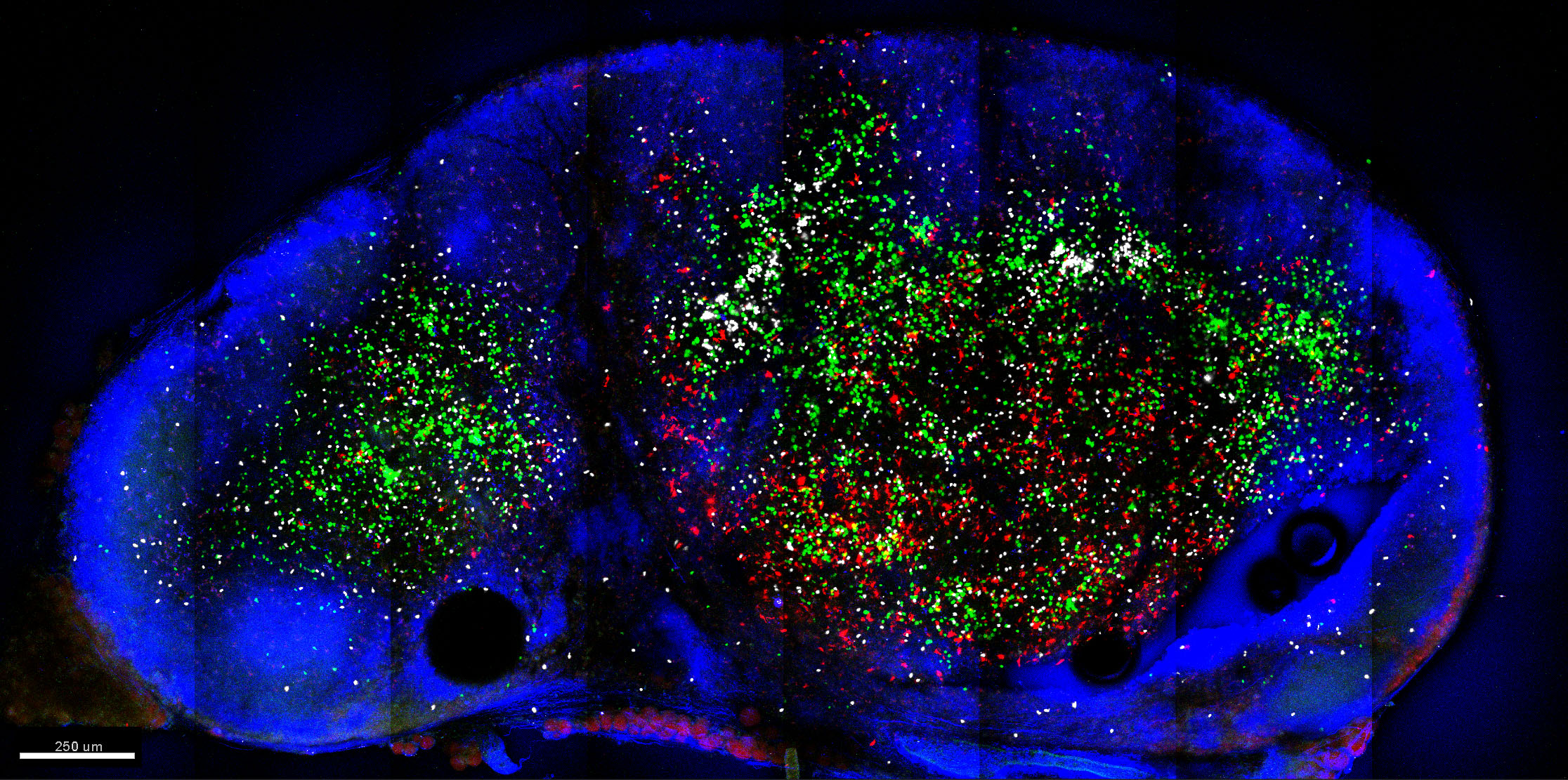
Figure 6. Example image of LN slice acquired using 2-photon microscopy. Shown is a 150 μm slice of mouse inguinal LN populated by CellTrace Violet-labelled CD4 T cells (green), CellTracker Deep Red-labelled CD8 T cells (white) and TRITC-labelled migratory DC (red). Cells were labelled with CellTrace or CellTracker dyes as per the manufacturer protocols (Thermo Fisher Scientific). LN sections were then stained with Pacific Blue anti-B220 (blue). Also in blue was collagen delineating the LN capsule illuminated via second-harmonic generation. Image was acquired using Zeiss LSM710 NLO at dual wavelengths (800 nm for excitation of CellTrace Violet, CellTracker Deep Red and Pacific Blue, and 880 nm for TRITC excitation) and at 512 x 512 resolution in 8 x 4 tiles for a 3 x 1.5 mm LN cross-section. Image was then processed using Imaris (Bitplane). Autofluorescence and spectral crossover were removed using Channel Arithmetic function in Imaris XTension.
Notes
- The best tissue quality can be obtained with slower cutting speed. We find that 0.03 mm/s slices the tissue with minimal disruption. However, if deemed acceptable, cutting speed can be increased to save time.
- Prolong Gold mounting media can also be used in conjunction with gelatin/glycerol to take advantage of their antifade properties: place small drops of Prolong Gold on top of tissue slices, and gelatin/glycerol mix in the empty area of the slide. This should allow Prolong Gold to cover the tissue slices while allowing the gelatin/glycerol mixture to effectively seal off the edges of the slide.
- Although a confocal microscope can be used to acquire images of vibratome slices (we have achieved up to 40 μm depth), it could not image deep enough to take full advantage of the slice’s thickness. A 2-photon microscope is preferable for imaging up to 150-200 μm depth.
- Depending on the fluorophore combination, dual 2-photon excitation wavelengths may be used to acquire fluorescence with distinct excitation cross-section (e.g., use 800 nm for AlexaFluor dyes and 920 nm for EGFP/DsRed).
- Certain fluorophores are rapidly bleached by two-photon excitation and should be carefully considered. For example, we found that antibody conjugated to AlexaFluor 647 to be a poor choice for such purpose. In contrast, AlexaFluor 594 is relatively photostable for two-photon imaging.
Recipes
- Periodate-Lysine-Paraformaldehyde fixative
For 10 ml solution:
3.75 ml 0.1 M P-buffer (pH 7.4)
3.75 ml 0.2 M L-lysine (pH 7.4)
0.0213 g NaIO4
2.5 ml 4% paraformaldehyde
Note: Prepared fresh for each use. - P-buffer, 0.1 M
For 200 ml solution:
19 ml 0.2 M NaH2PO4 (monobasic)
81 ml 0.2 M Na2HPO4 (dibasic)
100 ml H2O
Adjust pH to 7.4
Store at 4 °C - L-lysine, 0.2 M
For 100 ml solution:
2.92 g L-lysine
100 ml 0.1 M P-buffer
Adjust pH to 7.4
Store at 4 °C - 5% gelatin/glycerol mix
For 20 ml solution:
1 g gelatin from porcine skin
1 ml glycerol for molecular biology, ≥ 99%
20 ml PBS
Mix by inverting tube and leave in a 37 °C water bath to prevent hardening of gel
Note: Prepared fresh for each use.
Acknowledgments
This protocol was adapted from our publication (Hor et al., 2015). This work was supported by the National Health and Medical Research Council, and by the Australian Research Council.
References
- Hor, J. L., Whitney, P. G., Zaid, A., Brooks, A. G., Heath, W. R. and Mueller, S. N. (2015). Spatiotemporally distinct interactions with dendritic cell subsets facilitates CD4+ and CD8+ T cell activation to localized viral infection. Immunity 43(3): 554-565.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Hor, J. L. and Mueller, S. N. (2016). Imaging Thick Lymph Node Tissue Sections. Bio-protocol 6(18): e1938. DOI: 10.21769/BioProtoc.1938.
Category
Immunology > Immune cell imaging > Confocal microscopy
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










