- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Use of SCRI Renaissance 2200 (SR2200) as a Versatile Dye for Imaging of Developing Embryos, Whole Ovules, Pollen Tubes and Roots
Published: Vol 6, Iss 18, Sep 20, 2016 DOI: 10.21769/BioProtoc.1935 Views: 20372
Reviewed by: Samik BhattacharyaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Live Leaf-Section Imaging for Visualizing Intracellular Chloroplast Movement and Analyzing Cell–Cell Interactions
Yuta Kato [...] Mitsutaka Taniguchi
Aug 5, 2025 2322 Views

Live-Cell Monitoring of Piecemeal Chloroplast Autophagy
Masanori Izumi [...] Shinya Hagihara
Nov 5, 2025 1690 Views

Chloroplast Movement Imaging Under Different Light Regimes With a Hyperspectral Camera
Paweł Hermanowicz [...] Justyna Łabuz
Dec 20, 2025 756 Views
Abstract
Confocal laser scanning microscopy in combination with fluorescent proteins is a powerful tool for the study of sexual reproduction and other developmental processes in plants. In order to understand the origin and localization of fluorescent signals in a complex tissue, staining of cell outlines is often mandatory. Cell wall staining with SCRI Renaissance 2200 (SR2200) has recently been described as a method of choice to study plant reproductive processes (Musielak et al., 2015). In this protocol, we present detailed instructions on the use of SR2200 to stain cell walls in different Arabidopsis tissues.
Keywords: Renaissance SR2200Materials and Reagents
- Liquid blocker, “PAP pen” (Science Services, catalog number: N71310 )
- Microscope slides (e.g., Superfrost; Carl Roth, catalog number: 1879.1 )
- Double-sided adhesive tape (e.g., Reichelt Elektronik, Tesa Sande, catalog number: 05338 )
- Syringe needle (e.g., Terumo, catalog number: NN-2425R ; VWR International, catalog number: HSWA4710005525 )
- Microscope cover slips (e.g., Carl Roth, catalog number: H874.2 )
- Pipette tips (200 μl)
- Razor blade (e.g., VWR International, Darmstadt, catalog number: 233-5224 )
- Fresh Arabidopsis plant material (growth conditions as described in Babu et al., 2013)
- Deionized water
- 10% glycerol (e.g., Sigma-Aldrich, catalog number: 49782 )
- 1% (v/v) DMSO (e.g., Carl Roth, catalog number: 7029.2 )
- 0.05% (w/v) Triton-X 100 (e.g., Carl Roth, catalog number: 3051.2 )
- 3.75% (w/v) para-formaldehyde (e.g., Carl Roth, catalog number: 0335.2 )
- PBS buffer (pH 7.4) (e.g., Carl Roth, catalog number: 9143.2 )
- Optional: 4’,6-Diamidin-2-Phenylindol (DAPI) (e.g., Carl Roth, catalog number: 6335.1 )
- SR2200 staining solution (see Recipes)
Note: SR2200 can be ordered from: Renaissance Chemicals Ltd, Unit 1 Blackwood Hall Business Park, North Duffield, Selby, UK. Contact: Howard Weaver, enquiries@renchem.co.uk
Equipment
- Tweezers
- Binocular (e.g., ZEISS, model: Stemi 2000 )
- Adjustable pipette 20-200 μl (e.g., Gilson Pipetman-neo P200N; Carl Roth, catalog number: HY87.1 )
- Vacuum pump (e.g., Vacuubrand PC2004 Vario)
- Vacuum chamber (e.g., Kartell Labware 554)
- Confocal laser scanning microscope with 405 nm laser and standard filter set for DAPI imaging (e.g., Leica TCS SP8) or spectral detector if double staining with DAPI is required (e.g., Zeiss LSM780NLO with 32 channel GaAsP array)
Procedure
- Preparation and staining of Arabidopsis embryos (Figure 1)
- Using the PAP pen, mark a square on a microscope slide that frames a 20 mm by 20 mm area.
- Add 60 μl Renaissance staining solution to the center of the marked square.
- Remove a silique of the appropriate developmental stage with tweezers and transfer to double-sided tape on a microscope slide.
- Steps A5-8 will be carried out under a stereo microscope/binocular.
- Cut the silique open alongside the septum with a syringe needle (Video 1).Video 1. Dissecting siliques to collect ovules
- Gently pull the carpels to the side.
- Ovules can be picked up with the syringe needle tip and transferred to the staining solution on the microscope slide.
- Carefully cover the ovules with a cover slip avoiding air bubbles.
- Squeeze out the embryos from the seed by applying pressure on the cover slip with a pipette tip until ovules burst open. Slowly release the pressure to avoid turbulences that could break off the suspensor structure from the embryo (Video 2).
- Embryos can immediately be imaged by confocal microscopy.Video 2. Releasing embryos from dissected ovules by applying pressure to the coverslip
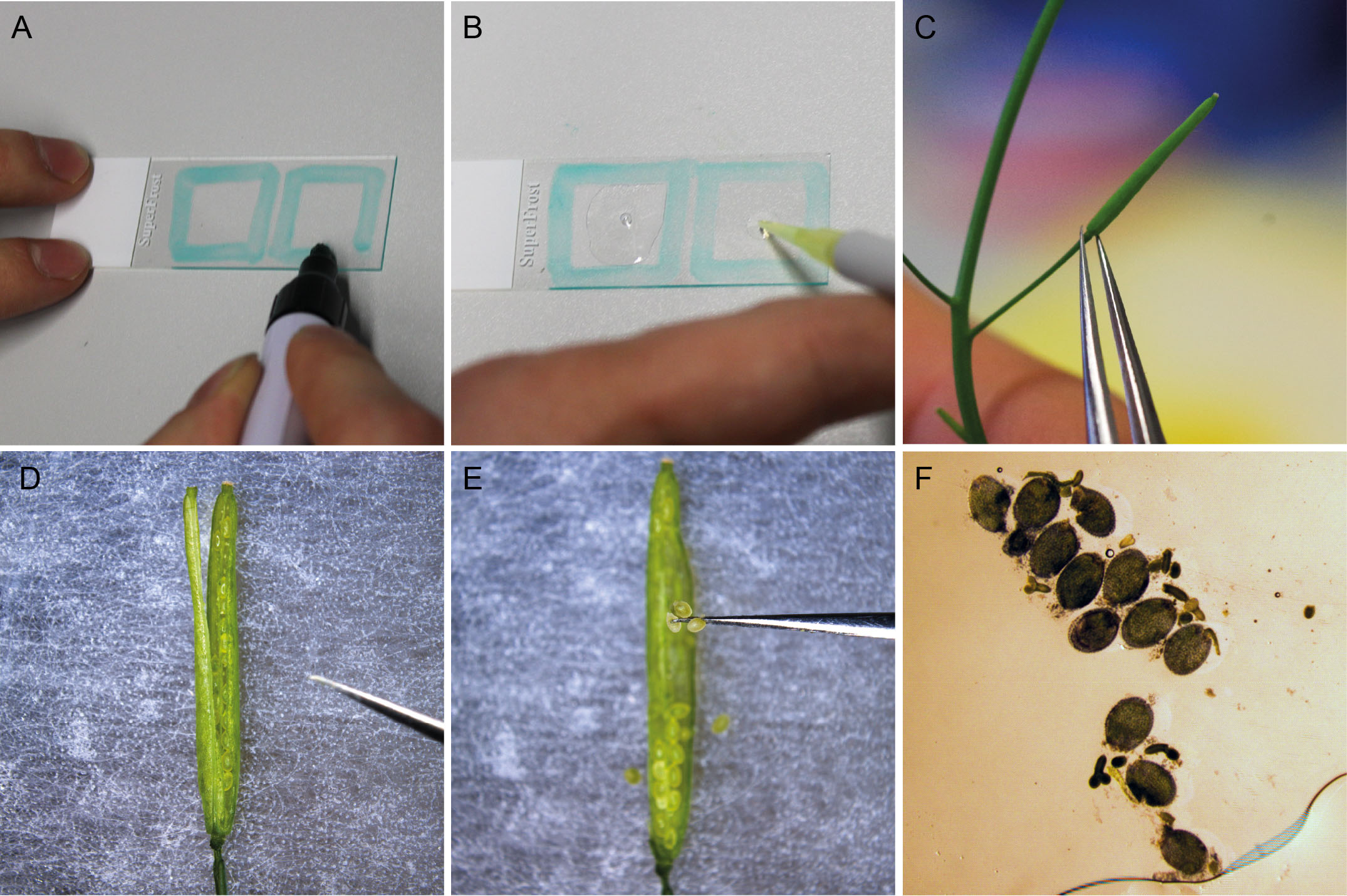
Figure 1. Preparation and staining of Arabidopsis embryos. A. Using the PAP pen, a 20 x 20 mm square is marked on a microscope slide. B. The square is filled with 60 µl SR2200 staining solution. C. A silique of the appropriate developmental stage is transferred to double-sided tape on a microscope slide. D. The silique is cut open using the tip of a syringe needle. E. Ovules are collected and transferred to the staining solution. F. After carefully covering the ovules with a cover slip, the embryos are squeezed out of the ovules by gently applying pressure to the cover slip with a pipet tip.
- Using the PAP pen, mark a square on a microscope slide that frames a 20 mm by 20 mm area.
- Preparation and staining of young ovules and pollen tubes
- Follow steps A1-7 of embryo preparation protocol.
- Unfertilized or freshly fertilized ovules (up to 2 days after pollination) as well as whole silique tissue can be transferred to the staining solution on the microscope slide. For young ovules, it is easiest to transfer the whole replum and septum with attached ovules.
- Carefully transfer the microscope slide to a vacuum chamber and apply soft vacuum (80 mbar) for 5 min.
- Remove the SR2200 staining solution with a pipette without picking up the stained plant tissue.
- Add 150 µl water.
- Apply soft vacuum (80 mbar) for 5 min.
- Carefully remove the water with a pipette without picking up the plant tissue.
- Add 50 μl 10% glycerol solution.
- Gently cover the sample with a cover slip without trapping air bubbles.
- The sample can immediately be used for confocal microscopy.
- Follow steps A1-7 of embryo preparation protocol.
- Preparation and staining of root tissue (Figure 2)
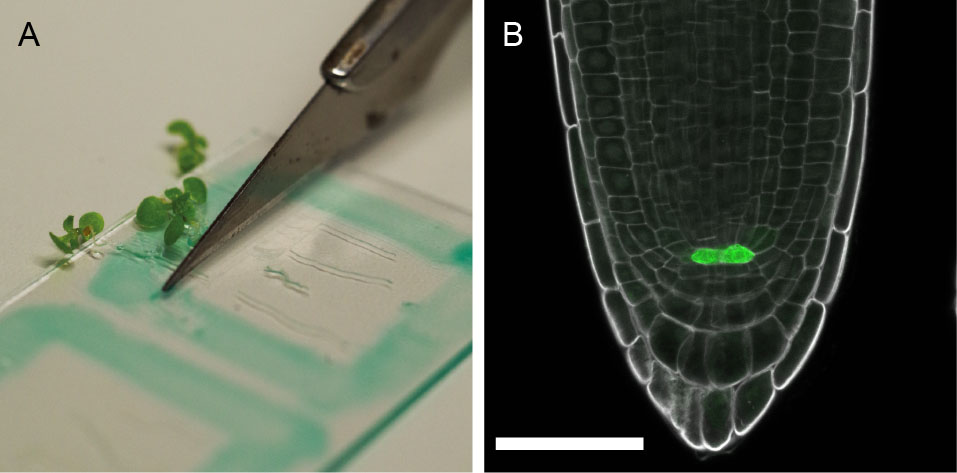
Figure 2. Preparation and staining of Arabidopsis roots. A. 5-day-old seedlings are placed on a prepared microscope slide and cut at the hypocotyl with a scalpel knife or razor blade before staining with SR2200 solution; B. pWOX5::erGFP expression (green) (Xu et al., 2006) in the quiescent center cells. SR2200 staining in grey. Scale bar = 50 µm.- Using the PAP pen, mark a square on a microscope slide that frames a 20 mm by 20 mm area.
- Transfer 4-5 seedlings to the object slide and position them within the square so that the roots are all orientated in one direction.
- Take a razor blade and cut horizontally below the hypocotyl.
- Add 100 μl SR2200 staining solution.
- Apply soft vacuum (80 mbar) for 5 min.
- Carefully remove the staining solution with a pipette without touching the roots.
- Add 100 µl water.
- Apply soft vacuum (80 mbar) for 5 min.
- Carefully remove the water with a pipette without touching the roots.
- Add 50 μl 10% glycerol solution.
- Gently cover the sample with a cover slip.
- The sample can immediately be used for confocal microscopy.
- Using the PAP pen, mark a square on a microscope slide that frames a 20 mm by 20 mm area.
- Microscopy
SR2200 has an excitation maximum around 350 nm but a 405 nm laser line can be used successfully. SR2200 gives a very bright fluorescence signal, therefore only very limited laser power is required. Emission can be detected using commonly available filter sets designed for DAPI imaging.
When double-staining with SR2200 and DAPI is desired, simultaneous imaging of both dyes in a single channel usually leads to a very weak DAPI signal in comparison to the much brighter SR2200 fluorescence. Therefore it is a more suitable approach to record spectral information of the fluorescence signal, for example with a detector array, and separate the signal into individual channels by spectral unmixing. We used a Zeiss LSM780 microscope with a 32-channel detector array in combination with the online fingerprinting function of the ZEN software and applied individually pre-recorded spectra of the two dyes as well as background fluorescence. The signal of the DAPI channel was increased around 18-fold in comparison to the signal of SR2200 by adjusting the digital gain of the individual channels to get comparable signal intensities. The staining procedure is as described above with the addition of DAPI to the staining solution.
Representative data
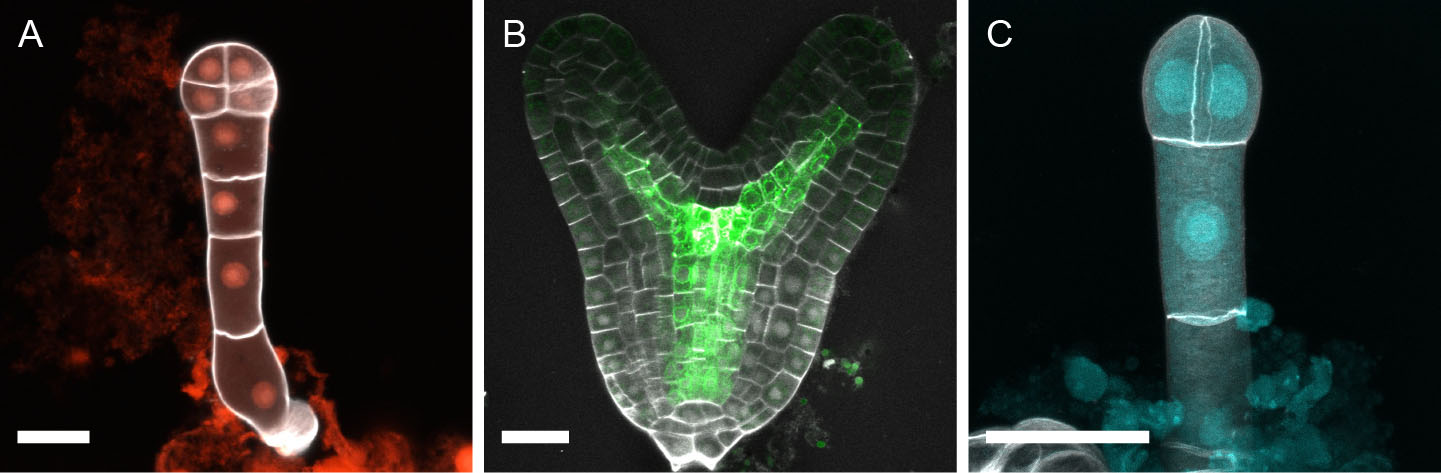
Figure 3. Representative images of SR2200-stained Arabidopsis embryos. A. 8-cell embryo expressing pNMA>>NLS-tdtomato (red) (Babu et al., 2013). B. Late heart-stage embryo expressing Q0990>>erGFP (green) (Weijers et al., 2006). SR2200 signal in grey. C. Maximum projection of 2-cell embryo double-stained with SR2200 and DAPI. DAPI signal in nuclei (cyan) was separated from SR2200 signal in cell walls (grey) by spectral unmixing (Musielak et al., 2015). Scale bar = 20 µm.
Recipes
- SR2200 staining solution
Note: Stock solution of the supplier is considered as 100%.
0.1% (v/v) SR2200
1% (v/v) DMSO
0.05% (w/v) Triton-X 100
5% (w/v) glycerol
3.75% (w/v) para-formaldehyde
in PBS buffer (pH 7.4)
Optional: 2 µg/ml DAPI
To simplify the preparation, we first prepare a 4% (w/v) para-formaldehyde (PFA) solution by dissolving the PFA powder in gently heated 1x PBS buffer (pH 7.4). After cooling of the PFA solution, we add the rest of the ingredients to make up the final volume. The staining solution can be stored in 1 ml aliquots at -20 °C. Always use fresh aliquots for staining.
Acknowledgments
This protocol was adapted from the previously published study, Musielak et al. (2015). We would like to thank Christian Liebig and Aurora Panzera of the microscope facility at the MPI for Developmental Biology for technical assistance and support, Agnes Henschen and Laura Schenkel for their help in establishing the initial staining protocol. Research in our group is supported by the German Science Foundation (Deutsche Forschungsgemeinschaft-DFG: SFB1101/B01 to M.B.) and the Max Planck Society.
References
- Babu, Y., Musielak, T., Henschen, A. and Bayer, M. (2013). Suspensor length determines developmental progression of the embryo in Arabidopsis. Plant Physiol 162(3): 1448-1458.
- Musielak, T. J., Schenkel, L., Kolb, M., Henschen, A. and Bayer, M. (2015). A simple and versatile cell wall staining protocol to study plant reproduction. Plant Reprod 28(3-4): 161-169.
- Weijers, D., Schlereth, A., Ehrismann, J. S., Schwank, G., Kientz, M. and Jurgens, G. (2006). Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev Cell 10(2): 265-270.
- Xu, J., Hofhuis, H., Heidstra, R., Sauer, M., Friml, J. and Scheres, B. (2006). A molecular framework for plant regeneration. Science 311(5759): 385-388.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Musielak, T. J., Bürgel, P., Kolb, M. and Bayer, M. (2016). Use of SCRI Renaissance 2200 (SR2200) as a Versatile Dye for Imaging of Developing Embryos, Whole Ovules, Pollen Tubes and Roots. Bio-protocol 6(18): e1935. DOI: 10.21769/BioProtoc.1935.
Category
Plant Science > Plant cell biology > Cell staining
Plant Science > Plant cell biology > Cell imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











