- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Peptide Loading on MHC Class I Molecules of Tumor Cells
Published: Vol 6, Iss 18, Sep 20, 2016 DOI: 10.21769/BioProtoc.1932 Views: 11602
Reviewed by: HongLok LungBenoit ChassaingAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Reprogramming Cancer Cells to Antigen-presenting Cells
Alexandra G. Ferreira [...] Carlos-Filipe Pereira
Nov 20, 2023 5005 Views

Isolation and Ex Vivo Testing of CD8+ T-Cell Division and Activation Using Mouse Splenocytes
Melissa Dolan [...] John M.L. Ebos
Aug 20, 2025 3878 Views
Abstract
MHC class I molecules present peptides to cytotoxic T cells allowing the immune system to scan for intracellular pathogens and mutated proteins. The generation of antigenic peptides is a multistep process that ends in the endoplasmic reticulum (ER). Only peptides with the right length and sequence will bind nascent MHC class I molecules in the ER. This protocol allows for detachment of the endogenous peptides bound to MHC class I molecules by preserving them for the binding of high affinity synthetic peptides. The complete dissociation of endogenous peptides by mild acid treatment as well as the binding of synthetic peptides to MHC class I molecules will be evaluated measuring HLA class I molecules express on the cell surface by flow cytometry. The mouse antibody W6/32 which recognizes β2m associated HLA-A, -B, -C, -E and -G heavy chains is suitable for this propose. Any tumor cell line that expresses surface HLA class I molecules is suitable for the assay. Another important aspect is to know the HLA class I typing of tumor cell line to allow selection of the known high affinity peptides.
Keywords: MHC class IMaterials and Reagents
- T25 Flasks (Corning, Falcon®, catalog numbers: 353108 )
- T75 Flasks (Corning, Falcon®, catalog numbers: 353136 )
- 15 ml centrifuge tubes (Corning, Falcon®, catalog number: 352096 )
- 6 well plates (Corning, Falcon®, catalog number: 353046 )
- 1.5 ml Safe-Lock tubes (Eppendorf, catalog number: 0030120086 )
- 96 well plates (Corning, catalog number: 3799 )
- FACS tubes
- Tumor cell line (grown in flasks, in incubator at 37 °C and 5% CO2)
- RPMI 1640 (Euroclone, catalog number: ECM9106L )
- Fetal bovine serum (FBS) heat-inactivated for 1 h at 56 °C (Thermo Fisher Scientific, Gibco®, catalog number: 10270-106 )
- Penicillin/streptomycin (EuroClone, catalog number: ECB3001D )
- L-glutamine (EuroClone, catalog number: ECB3000D )
- EDTA (Sigma-Aldrich, catalog number: E5134 )
- Phosphate buffered saline (PBS) (EuroClone, catalog number: ECB4004L )
- Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A2153-100G )
- Trypan-Blue
- Na2HPO4 (Sigma-Aldrich, catalog number: S5136 )
- Citric acid monohydrate (Sigma-Aldrich, catalog number: 251275 )
- mAb W6/32
- Beta 2-microglobulin (Sigma-Aldrich, catalog number: M4890 )
- Peptides (lyophilized, resuspended in DMSO at 10 mM) (Anaspec)
- Goat F(ab’)2 Fragment anti-mouse IgG (H+L)-FITC (Beckman Coulter, catalog number: PN IM0819 )
- Complete RPMI (see Recipes)
- 833 μM EDTA (see Recipes)
- FACS buffer (see Recipes)
- Acid buffer (see Recipes)
Equipment
- Centrifuge, used with maximal acceleration and deceleration (Eppendorf, model: 5810R )
- Incubator (5% CO2, 37 °C) (FormaTM Steri-CultTM CO2 Incubator-Thermo Fisher Scientific)
- Flow cytometer equipped with 2 lasers (8 detectors), interfaced with PC using DIVA Software version 6.1.3 (BD, model: FACSCanto II )
Software
- DIVA software version 6.1.3
- FlowJo software
Procedure
- Acid treatment of tumor cell lines
- Tumor cells are cultured in fresh RPMI medium at a cell density ≥ 0.5-2 x 106 cell/ml depending on the type of cells, in an incubator at 37 °C, 5% CO2.
- Wash adherent cells in PBS (5 ml) and detach cells by adding 833 μM EDTA (1 ml for T25 flask and 2 ml for T75 flask) for 3-5 min in an incubator at 37 °C, 5% CO2.
- Wash cells twice with PBS (2 x 5 ml in 15 ml tube) by centrifugation at 468 x g for 7 min and re-suspend them in cold FACS buffer.
- Count total viable cells with Trypan-Blue exclusion method.
- Harvest 5-10 x 106 cells with 5 ml PBS (at RT) in 15 ml tubes.
- Centrifuge cells at 468 x g for 7 min.
- Discard the supernatant and re-suspend the cells in 600 μl cold acid buffer by manual gentle shaking.
- Incubate for 1 to 3 min on ice (preliminary experiments should be performed for each cell line to detect the best condition to completely dissociate MHC class I complexes by preserving cell viability; the complete dissociation will be deduced with the cell surface expression of HLA class I molecules detected by staining the cells with the mAb W6/32).
- Immediately add 10 ml of complete RPMI (at RT).
- Centrifuge cells at 468 x g for 7 min.
- Discard supernatant and re-suspend the cells in 10 ml of complete RPMI.
- Centrifuge cells at 468 x g for 7 min.
- Discard supernatant and re-suspend the cells in 1 ml of complete RPMI containing beta 2-microglobulin (5 μg/ml).
- Tumor cells are cultured in fresh RPMI medium at a cell density ≥ 0.5-2 x 106 cell/ml depending on the type of cells, in an incubator at 37 °C, 5% CO2.
- Pulsing with peptides
- Seed the cells in 6 well plate at a cell density of 1 x 106 cells/well. Prepare 1 well for the control (without peptide) and 1 well for each peptide to be tested and/or a mix of different peptides.
- Add 50 μM of peptide (stocked at 10 mM, single or a mix of different peptides).
- Incubate for 2 h at RT and for additional 2 h at 37 °C in an incubator.
- Seed the cells in 6 well plate at a cell density of 1 x 106 cells/well. Prepare 1 well for the control (without peptide) and 1 well for each peptide to be tested and/or a mix of different peptides.
- Staining and FACS analysis
- Harvest cells in 1.5 ml Safe-Lock tubes and centrifuge at 468 x g for 7 min.
- Discard the supernatant and re-suspend the cells in 100 μl of cold FACS buffer.
- Distribute in a round-bottom 96 well plate (50 μl/well).
- Centrifuge the plate at 832 x g for 2 min at 4 °C.
- Discard the supernatant and re-suspend cells in 50 μl of cold FACS buffer containing mAb W6/32 (diluted 1:500) or in 50 μl of cold FACS buffer without mAb; mix carefully.
- Incubate for 25 min on ice.
- Add 150 μl of cold FACS buffer and centrifuge at 832 x g at 4 °C.
- Discard the supernatant and re-suspend cells in 50 μl of cold FACS buffer containing goat F(ab’)2 Fragment anti-mouse IgG (H+L)-FITC (diluted 1:100); mix carefully.
- Incubate for 25 min on ice in the dark.
- Add 150 μl of cold FACS buffer and centrifuge at 832 x g at 4 °C.
- Discard the supernatant and re-suspended cells in 150 µl of cold FACS buffer in FACS tubes and perform flow cytometric analysis. Viable human single cells will be selected by gating in forward scatter versus side scatter dot plot and the amount of cell surface MHC class I expression will be evaluated by mean fluorescence intensity (mfi) (Figure 1). Pulsing with high affinity peptides allows the recovery of surface MHC class I expression of acid-treated cells.
- Acquire data using DIVA software and analyse using FlowJo software.
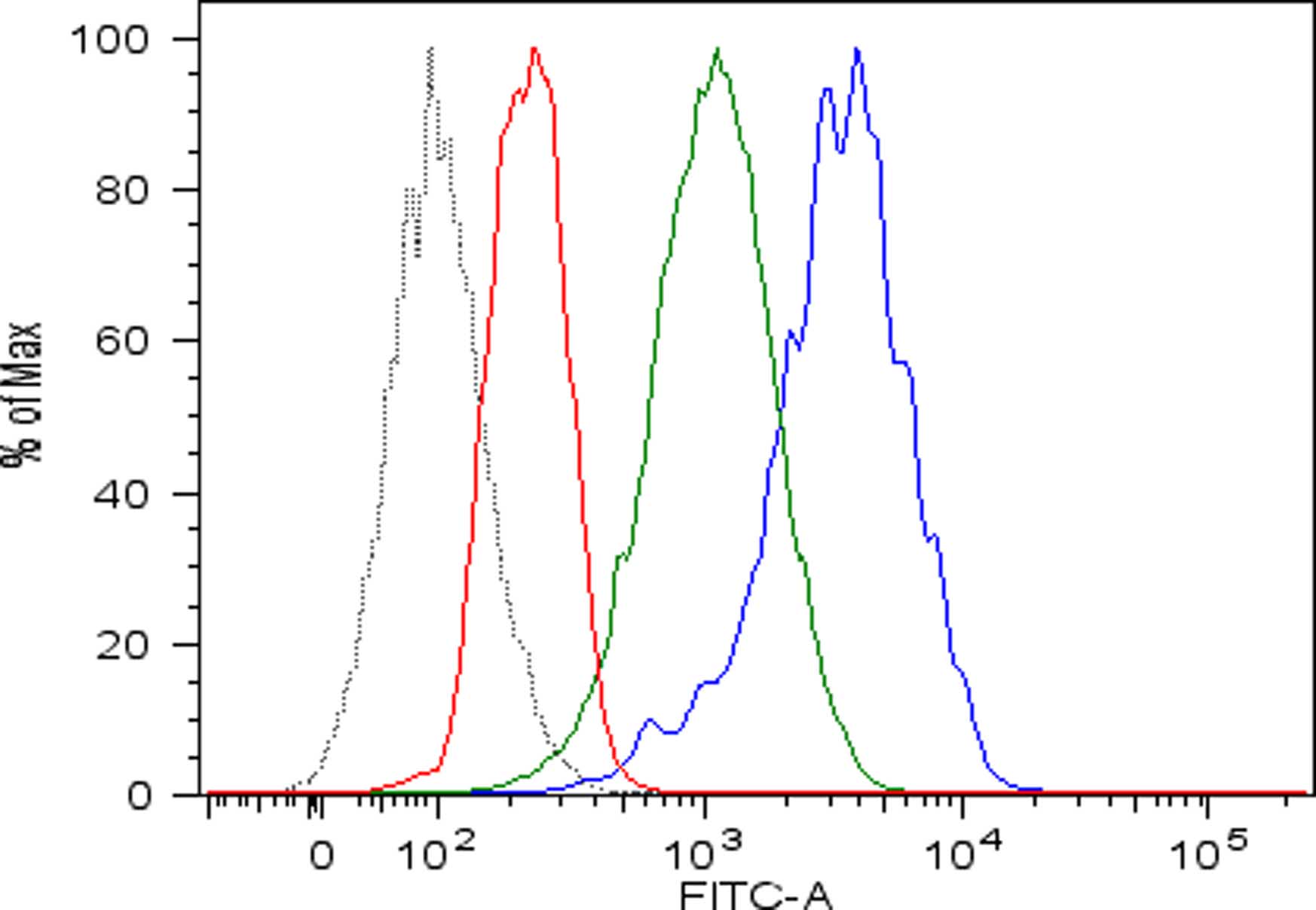
Figure 1. Recovery of peptide-MHC class I complexes on the surface of acid-treated DAOY cells pulsed with high affinity peptides. Flow cytometric analysis of MHC class I expressed on DAOY tumor cells left untreated (blue line) or pretreated with low pH buffer to allow dissociation of preformed peptide-MHC class I complexes (red line) and then pulsed with high affinity peptides (green line). Isotype-matched negative control antibody displayed as the black line.
- Harvest cells in 1.5 ml Safe-Lock tubes and centrifuge at 468 x g for 7 min.
Recipes
- Complete RPMI
440 ml RPMI 1640
50 ml FBS
5 ml penicillin-streptomycin
5 ml L-glutamine - 833 μM EDTA
Dissolve 7.3 g EDTA in 50 ml sterile water
Adjust pH to 8.0
Add 83.3 μl of this solution in 50 ml sterile PBS
Filter sterilize through a 20 µm filter
Store at 4 °C - FACS buffer
50 ml PBS
1 g BSA
4.Acid buffer
Dissolve 2.52 g Na2HPO4 (263 mM) and 1.29 g Citric acid monohydrate (123 mM) in 50 ml sterile water
Adjusted pH to 2.5
Acknowledgments
This protocol was adapted from Cifaldi et al. (2015). We acknowledge funding from Italian Ministry of Health (Rome, Italy) grant and the special project 5 x 1,000 Associazione Italiana per la Ricerca sul Cancro (AIRC, Milan, Italy) grant.
References
- Cifaldi, L., Romania, P., Falco, M., Lorenzi, S., Meazza, R., Petrini, S., Andreani, M., Pende, D., Locatelli, F. and Fruci, D. (2015). ERAP1 regulates natural killer cell function by controlling the engagement of inhibitory receptors. Cancer Res 75(5): 824-834.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Cifaldi, L., Locatelli, F. and Fruci, D. (2016). Peptide Loading on MHC Class I Molecules of Tumor Cells. Bio-protocol 6(18): e1932. DOI: 10.21769/BioProtoc.1932.
Category
Immunology > Immune cell function > Antigen-specific response
Cancer Biology > Tumor immunology > Cell biology assays
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










