- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Identification of RNA-binding Proteins
Published: Vol 6, Iss 17, Sep 5, 2016 DOI: 10.21769/BioProtoc.1920 Views: 14679
Reviewed by: Antoine de MorreeZhen ShiKate Hannan

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

ZnCl2 Precipitation-Assisted Sample Preparation for Proteomic Analysis
Qiqing He [...] Fuchu He
Jul 20, 2025 2733 Views

Protocol for the Preparation of a Recombinant Treacle Fragment for Liquid–Liquid Phase Separation (LLPS) Assays
Nadezhda V. Petrova [...] Artem K. Velichko
Sep 20, 2025 1823 Views

Optimized Secretome Sample Preparation From High Volume Cell Culture Media for LC–MS/MS Proteomic Analysis
Basil Baby Mattamana [...] Peter Allen Faull
Dec 20, 2025 1261 Views
Abstract
This protocol describes the extraction of RNA-binding proteins (RBPs) from cell lysates. In order to pull down target RBPs, 5-bromo-UTP (BrUTP)-incorporated RNA probes are used, which are generated by in vitro transcription. The schematic diagram (Flowchart) with procedure is indicated (Figure1 and Figure 2).
Figure 1. Schematic diagram of procedure (A-H). Flow chart of experimental procedure is indicated at A-H.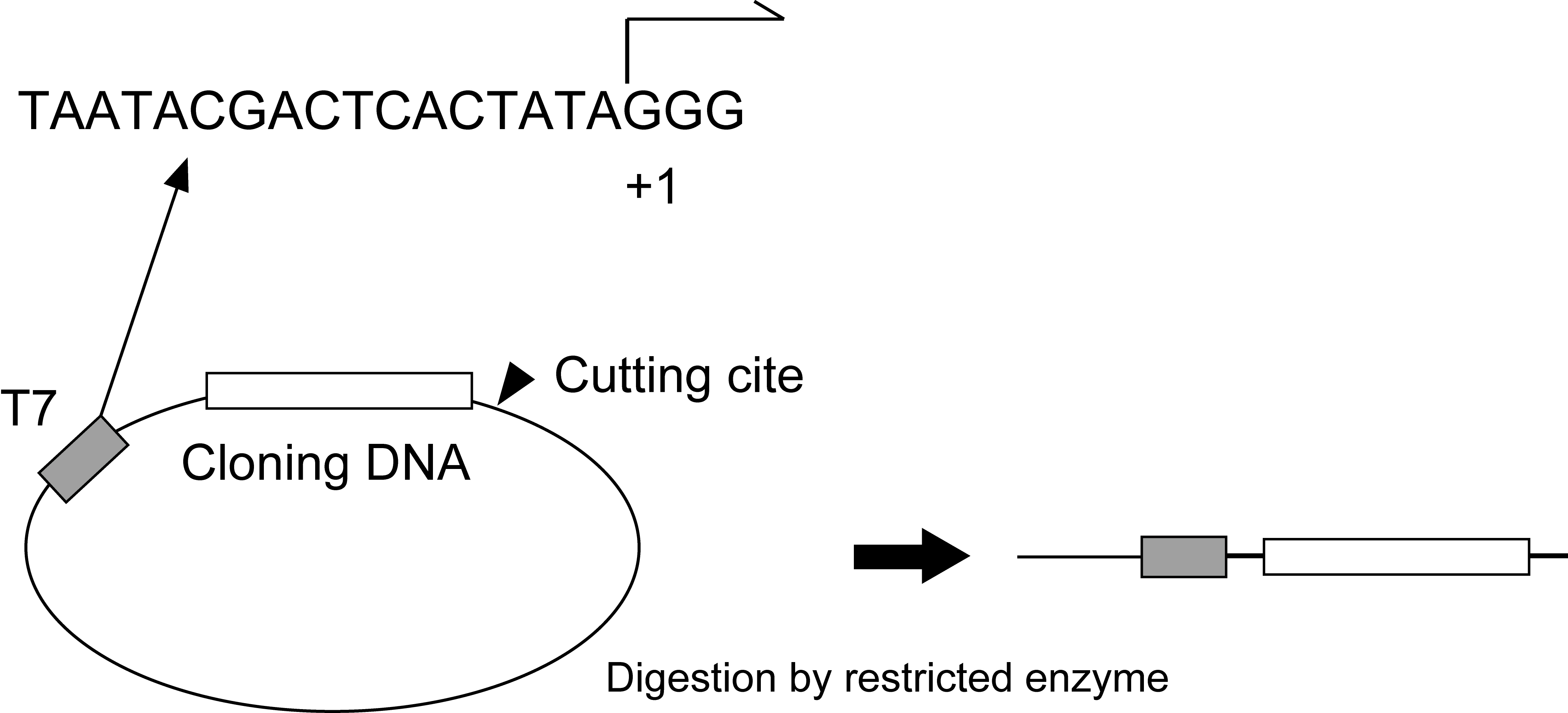
Figure 2. Linearization of plasmids by restricted enzyme. The plasmid is cut at restriction sites adjacent to its cloning element.
Materials and Reagents
- Microcentrifuge tubes (1.5 to 2.0 ml)
- Desalting MobiSpin columns (MoBiTec, catalog number: M105035F )
- DNA template (e.g., pBluescript vector encoding the target sequences such as non-coding elements IL-6 3’UTR and TNF-α 3’UTR)
- Reagents for in vitro transcription kits (TAKARA BIO, catalog number: 6140 )
- 5-bromouridine 5’-triphosphate (sodium salt) (Cayman Chemical, catalog number: 18140 )
- 50 mM ATP solution
- 50 mM GTP solution
- 50 mM CTP solution
- 50 mM UTP solution
- RNaseOUTTM recombinant ribonuclease inhibitor (Thermo Fisher Scientific, InvitrogenTM, catalog number: 10777-019 )
- T7 RNA polymerase
- DNase I (RNase free) (New England Biolabs, catalog number: M0303S )
- Protease inhibitor cocktail (Sigma-Aldrich, catalog number: MSSAFE )
- Protease inhibitor cocktail (NACALAI TESQUE, catalog number: 0 4080 )
- TRIzol® Reagent (Thermo Fisher Scientific, AmbionTM, catalog number: 15596-026 )
- Anti-BrdU antibody (IIB5) (Abcam, catalog number: ab8152 )
- Protein G Sepharose 4 fast flow (GE Healthcare, catalog number: 17061801 )
- Nuclease-free PBS (NACALAI TESQUE, catalog number: 14249 )
- Nuclease-free water (Thermo Fisher Scientific, AmbionTM, catalog number: 4387936 )
- Dithiothreitol (DTT) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: R0861 )
- Chloroform (NACALAI TESQUE, catalog number: 08401-65 )
- DEPC water (Thermo Fisher Scientific, AmbionTM, catalog number: AM9916 )
- 96% ethanol or 70% ethanol (NACALAI TESQUE, catalog number: 09666-85 )
- NE-PER nuclear and cytoplasmic extraction reagents (Thermo Fisher Scientific, catalog number: 78833 )
- 0.25% trypsin-EDTA solution (Thermo Fisher Scientific, catalog number: 25200056 )
- 1 M Tris-HCl, pH 7.4 (NACALAI TESQUE, Gibco®, catalog number: 35436-01 )
- 5 M NaCl (NACALAI TESQUE, catalog number: 31320-05 )
- 0.1 M spermidine (Sigma-Aldrich, catalog number: S2626-1G )
- 10% Nonidet(R) P-40 (NACALAI TESQUE, catalog number: 25223-04 )
- 0.5 M EDTA (NACALAI TESQUE, catalog number: 06894-14 )
- 1 M MgCl2 (NACALAI TESQUE, catalog number: 20942-34 )
- 1% Tween 20 (NACALAI TESQUE, catalog number: 35624-15 )
- PBS, pH 7.0 (NACALAI TESQUE, catalog number: 14249-24 )
- 1% SDS (NACALAI TESQUE, catalog number: 30562-04 )
- Coomassie brilliant blue (CBB) (Thermo Fisher Scientific, catalog number: 20278 )
- 10x transcription buffer (see Recipes)
- Bead washing buffer (see Recipes)
- RNA-binding buffer (see Recipes)
- RNA-protein wash buffer (see Recipes)
- Lysis buffer (see Recipes)
- Elution buffer (see Recipes)
Equipment
- MALDI-QIT-TOF (Shimadzu Europa GmbH, model: AXIMA Resonance )
- Microcentrifuge capable of reaching up to 16,000 x g
- Vortex mixer
- Centrifuge capable of reaching up to 2,000 x g
Procedure
- In vitro transcription of 5-bromo-UTP (BrUTP)-incorporated RNA probes
Note: DNA template such as pBluescript vector encoding the target sequences should be made linear DNA by restriction enzyme digestion and purified by phenol/chloroform extraction and ethanol precipitation (Figure 2).- Preparation of DNA template such as plasmids, PCR-generated or synthetic oligonucleotides.
- In vitro transcription reaction:
20 ng-1 μg DNA template
2 μl 10x transcription buffer
2 μl 50 mM ATP solution
2 μl 50 mM GTP solution
2 μl 50 mM CTP solution
1 μl 50 mM UTP solution
1 μl 50 mM 5-bromo-UTP (BrUTP) solution
0.5 μl RNase inhibitor
2 μl T7 RNA polymerase
X μl RNase free ddH2O (up to total 20 μl)
Mix by pipetting, and centrifuge. - Incubate for 2 h at 42 °C.
- Add 10-20 U DNase to 20 μl total solution to digest any DNA. After mixing, incubate for 1-2 h at 37 °C.
- The RNA probes are purified using Trizol following standard protocol.
Optional: If you want to confirm the size of RNA transcripts, agarose gel electrophoresis of RNA in formaldehyde will be performed following standard protocol.
- Preparation of DNA template such as plasmids, PCR-generated or synthetic oligonucleotides.
- Conjugation of anti-BrdU antibodies with protein G beads
- Wash protein G agarose beads three times with an equal volume of nuclease-free PBS (centrifuge at 2,000 x g for 1 min at 4 °C).
- Apply 100 μl of the solution (50% beads made up with PBS) to 1.5 ml new microcentrifuge tubes.
- Add 500 μl of bead wash buffer to each tube.
- Add 50 μl of anti-BrUTP m Ab to each tube.
- Incubate the tubes while rotating for at least 1 h at 4 °C. The sample can be incubated at 4 °C overnight.
- Wash antibody-conjugated beads once with 1 ml of bead wash buffer (centrifuge at 2,000 x g for 1 min at 4 °C).
- Discard the supernatants carefully.
- Wash protein G agarose beads three times with an equal volume of nuclease-free PBS (centrifuge at 2,000 x g for 1 min at 4 °C).
- Binding of BrUTP-labeled RNA to antibody-conjugated beads
- Resuspend the antibody-conjugated beads with 500 μl of bead wash buffer.
- Add 50 pmol of prepared BrUTP-labeled RNA (prepared in step A), and RNase inhibitor.
- Incubate while rotating for 2 to 3 h at 4 °C.
- Centrifuge at 2,000 x g for 1 min at 4 °C.
- Discard supernatant and wash BrUTP labeled RNA bound to the antibody-conjugated beads with 500 μl of bead wash buffer (centrifuge at 2,000 x g for 1 min at 4 °C) (To step E).
- Resuspend the antibody-conjugated beads with 500 μl of bead wash buffer.
- Pre-clearing the protein extraction
Optional: Cytoplasmic and nuclear protein extract can be separated using NE-PER Nuclear and cytoplasmic extraction reagents. Protein extracts were prepared from at least 1.0 x 108 cells.- The preparation of the protein extracts
- Collect the cultured cells into a 1.5 ml new microcentrifuge tube (if dissociation is necessary, Trypsin-EDTA solution can be used).
- Centrifuge the tube at 2,000 x g for 3 min at 4 °C.
- Discard the supernatants carefully.
- Transfer 1 ml lysis buffer into the tube with cell pellet, and resuspend the cells on ice.
- Put the tube on ice for 10 min.
- Centrifuge the tube at 16,000 x g for 10 min at 4 °C.
- Transfer the supernatant (protein extract) carefully into a 1.5 ml new microcentrifuge tube.
- Store 50 μl protein extract as input.
- Collect the cultured cells into a 1.5 ml new microcentrifuge tube (if dissociation is necessary, Trypsin-EDTA solution can be used).
- The preparation of protein G beads
- Wash 50 μl protein G agarose beads three times with an equal volume of nuclease-free PBS (centrifuge at 2,000 x g for 1 min at 4 °C). Discard supernatant gently.
- Apply 100 μl of the solution (50% beads made up with PBS) to 1.5 ml new microcentrifuge tubes (50 μl beads per sample).
- Add 500 μl of bead wash buffer to each tube.
- Centrifuge the tubes at 2,000 x g for 1 min at 4 °C.
- Discard the supernatant carefully.
- Wash 50 μl protein G agarose beads three times with an equal volume of nuclease-free PBS (centrifuge at 2,000 x g for 1 min at 4 °C). Discard supernatant gently.
- The mixture of the protein extract and 50% bead solution
- Transfer the protein extract into the tube with protein G beads.
- Incubate the tube while rotating for 1 h at 4 °C.
- Transfer the protein extract into the tube with protein G beads.
- The preparation of the protein extracts
- Binding of proteins to BrUTP-labeled RNA-conjugated beads
- Centrifuge the sample tubes containing the protein extract and protein G agarose beads at 2,000 x g for 2 min at 4 °C (save 10 μl of the protein extract as an input).
- Transfer the protein extract (optional: either purified cytoplasmic or nuclear protein extract) into the tubes prepared in step C.
- Incubate with rotation for 2 h at 4 °C.
- Centrifuge the sample tubes containing the protein extract and protein G agarose beads at 2,000 x g for 2 min at 4 °C (save 10 μl of the protein extract as an input).
- Purification of the binding proteins to RNA-conjugated beads
- After incubation, centrifuge the samples at 2,000 x g for 1 min at 4 °C. Discard the supernatant carefully.
- The protein-RNA complex on the beads is washed three to four times with 1 ml RNA-binding buffer. Centrifuge at 2,000 x g for 2 min at 4 °C.
- After incubation, centrifuge the samples at 2,000 x g for 1 min at 4 °C. Discard the supernatant carefully.
- Elution of proteins binding to BrUTP-labeled RNA-conjugated beads
- Resuspend the beads in 200 μl of nuclease-free PBS and transfer the slurry to a bottom-plugged spin column.
- Detach the bottom plug from the spin column, and then put the column into a new centrifuge tube. Centrifuge the column at 1,000 x g for 30 sec at 4 °C.
- Attach the bottom plug to the spin column, and put the column into a new centrifuge tube. Add 50 μl of elution buffer into the column.
- Incubate for 30 min at 4 °C with gentle shaking.
- Detach the bottom plug from the spin column, and put the column into a new centrifuge tube.
- Elute the protein-BrUTP-labeled RNA complexes by centrifugation.
- For enrichment of purified proteins, repeat elution steps G3-6, and collect the eluates into a new tube.
- Resuspend the beads in 200 μl of nuclease-free PBS and transfer the slurry to a bottom-plugged spin column.
- Detection of RNA-binding proteins by LC/MS/MS
- The eluted samples are subjected to SDS-PAGE, followed by CBB staining.
- Cut the gel on the several compartments, in which some bands were detected.
- The samples cut are eluted, and then analyzed by LC/MS/MS.
- Target RNA-binding proteins are confirmed by Western blotting (After step G).
- The eluted samples are subjected to SDS-PAGE, followed by CBB staining.
Recipes
- 10x transcription buffer
0.4 M Tris-HCl, pH 7.4
100 mM MgCl2
0.5 M NaCl
0.1 M spermidine - Bead washing buffer
20 mM Tris-HCl, pH 7.4
137 mM NaCl
1% NP-40
2 mM EDTA
1.5 mM DTT - RNA-binding buffer
0.2 M Tris-HCl, pH 7.4
0.5 M NaCl
20 mM MgCl2
1% Tween 20 - RNA-protein wash buffer
20 mM Tris-HCl, pH 7.4
10 mM NaCl
1% Tween 20 - Lysis buffer
50 mM Tris-HCl, pH 7.4
150 mM NaCl
1% NP-40
1% protease inhibitor cocktail
1 mM DTT - Elution buffer
PBS (pH 7.0)
1% SDS
Acknowledgments
This work was supported by funds of the Japanese Science and Technology Agency and the Japanese Ministry of Education, Culture, Sports, Science and Technology for integrated promotion of social system reform and research and development, and the Kishimoto foundation.
References
- Masuda, K., Ripley, B., Nishimura, R., Mino, T., Takeuchi, O., Shioi, G., Kiyonari, H. and Kishimoto, T. (2013). Arid5a controls IL-6 mRNA stability, which contributes to elevation of IL-6 level in vivo. Proc Natl Acad Sci U S A 110(23): 9409-9414.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Masuda, K. and Kishimoto, T. (2016). Identification of RNA-binding Proteins. Bio-protocol 6(17): e1920. DOI: 10.21769/BioProtoc.1920.
Category
Biochemistry > RNA > RNA-protein interaction
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









