- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Measurements of Proline and Malondialdehyde Content and Antioxidant Enzyme Activities in Leaves of Drought Stressed Cotton
Published: Vol 6, Iss 17, Sep 5, 2016 DOI: 10.21769/BioProtoc.1913 Views: 30485
Reviewed by: Tie LiuHideyuki TakahashiZhanwu Dai

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Simple Sonication Method to Isolate the Chloroplast Lumen in Arabidopsis thaliana
Jingfang Hao and Alizée Malnoë
Aug 5, 2023 2284 Views

A Plate Growth Assay to Quantify Embryonic Root Development of Zea mays
Jason T. Roberts [...] David M. Braun
Oct 20, 2023 2255 Views

Detection and Quantification of Programmed Cell Death in Chlamydomonas reinhardtii: The Example of S-Nitrosoglutathione
Lou Lambert and Antoine Danon
Aug 5, 2024 1588 Views
Abstract
Drought stress negatively affects cotton plant growth and induces various biochemical and physiological responses in cotton plants. Proline content and antioxidant enzymes are thought to be associated with maintaining the structure of cellular components or with protecting cellular function. Study of cotton plant responses towards drought stress and investigation of the mechanism of drought tolerance are helpful to develop drought tolerant cotton plants. Here, we describe a protocol to investigate cotton plant response towards drought stress through measurements of biochemical parameters including antioxidant enzyme activities, proline content and malondialdehyde (MDA) content.
Materials and Reagents
- Centrifuge tubes (1.5 ml) (Corning, Axygen®, catalog number: MCT-150-C-S )
- Gossypium barbadense seeds
- Soil mix (vermiculite:humus = 1:1) (http://www.xing-nong.cn/ProductList.Asp?SortID=207)
- 1/2 MS media salt without sugar and agar (Hopebio, catalog number: HB8469-12 )
- Sodium phosphate dibasic anhydrous (Na2HPO4) (Sinopharm Chemical Reagent, catalog number: 20040618 )
- Sodium phosphate monobasic anhydrous (NaH2PO4) (Sinopharm Chemical Reagent, catalog number: 20040818 )
- ddH2O
- Liquid nitrogen
- Ice
- Ninhydrin (C9H4O3·H2O) (Sinopharm Chemical Reagent, catalog number: 30130212 )
- Trichloroacetic acid (TCA) (Sinopharm Chemical Reagent, catalog number: 80132618 )
- 5-sulphosalicylic acid dihydrate (Sinopharm Chemical Reagent, catalog number: 10021516 )
- L-proline (Genview, catalog number: FL259 )
- Thiobarbituric acid (TBA) (Sigma-Aldrich, catalog number: T5500 )
- Sodium hydroxide (NaOH) (Sinopharm Chemical Reagent, catalog number: 10019762 )
- EDTA-2Na (Sigma-Aldrich, catalog number: 27285 )
- L-methionine (Sigma-Aldrich, catalog number: V900487 )
- Nitroblue tetrazolium (NBT) (Beijing Dingguo Changsheng Biotechnology, catalog number: JN213 )
- Riboflavin (Sigma-Aldrich, catalog number: 47861 )
- Guaiacol (Sigma-Aldrich, catalog number: G5502 )
- 30% hydrogen peroxide (H2O2) (Sinopharm Chemical Reagent, catalog number: 10011208 )
- Ethanol (Sinopharm Chemical Reagent, catalog number: 10009259 )
- Glutathione reduced (GSH) (Sigma-Aldrich, catalog number: G4251 )
- 1-chloro-2, 4-dinitrobenzene (CDNB) (Sigma-Aldrich, catalog number: C38702 )
- Acetic acid (Sinopharm Chemical Reagent, catalog number: 10000208 )
- 1/2 MS solution (see Recipes)
- Na2HPO4 stock solution (see Recipes)
- NaH2PO4 stock solution (see Recipes)
- PBS (pH 7.0) (see Recipes)
- 2.5% acid-ninhydrin (see Recipes)
- 3% sulphosalicylic acid (see Recipes)
- 10% TCA (see Recipes)
- 0.25% TBA (see Recipes)
- PBS (pH 7.8) (see Recipes)
- 1 mM EDTA-2Na (see Recipes)
- 130 mM methionine (see Recipes)
- 750 μM NBT (see Recipes)
- 20 μM riboflavin (see Recipes)
- 0.2% guaiacol (see Recipes)
- PBS (pH 6.5) (see Recipes)
- 5 mM GSH (see Recipes)
- 1.5 mM CDNB (see Recipes)
Equipment
- Plant growth chamber (25 °C, 16 h/8 h photoperiod, 10,000 lux, 50-60% humidity)
- Mortar and pestle
- Plastic pots (8 cm diameter and 12 cm depth) (http://www.lyyyb.com/yingyangbo.html)
- Electronic balance (Sartorius, model: BAS124S-CW )
- ddH2O purification system (Xiamen RSJ Scientific Instruments, model: Spring-S10 )
- Ice making machine (Xueke, model: IMS-40 )
- Centrifuge (Eppendorf, model: 5810 R )
- Spectrophotometer (Analytik Jena, model: ScanDrop® 250)
- Water bath (Changzhou Aohua Instrument, model: HH-1 )
- Pipette, 2-20 µl (Eppendorf, Eppendorf Research® plus, model: 3120000038 )
- Pipette, 20-200 µl (Eppendorf, Eppendorf Research® plus, model: 3120000054 )
- Pipette, 100-1,000 µl (Eppendorf, Eppendorf Research® plus, model: 3120000062 )
Procedure
- Soak cotton seeds in water overnight at 28 °C to promote seed germination.
Note: Seeds germinate after 24 h. - Next day, sow germinated seeds into pots filled with soil mix (vermiculite:humus = 1:1), one seed per pot.
Note: Pots with small holes at the bottom are important to leak surplus water. - Maintain plantlets in a plant growth chamber with controlled conditions of 25 °C and 16 h light/8 h dark photoperiod at a light intensity of 10,000 lux during the experiment. Regularly water the pots with liquid 1/2 MS media (300 ml per pot) to provide nutrition to the plants.
Note: The regular supplement of 1/2 MS media in the soil is required for plant growth in pots. After 7~10 days for two cotyledons to fully expand under the controlled conditions, this is a suitable time to perform the infiltration operation of virus-induced gene silencing (VIGS) assay in cotton (Chen et al., 2015; Gao et al., 2011; Gu et al., 2014). - After 3-4 weeks, pots with two-three-leaves plantlets are watered every day for a period of seven days to saturation.
Note: Surplus water could leak out from holes at the bottom, and a condition of saturated moisture in the soil provides similar water content before drought treatment. - For drought treatment, plantlets are then deprived of water for 35 days, followed by re-watering once. Seven days after the re-watering treatment, cotton plants with at least one un-wilting green young leaf as well as a living shoot apex is regarded as “survival” (Figure 1A). Survival rate is calculated as the ratio of number of survived plants over the total number of treated plants (Figure 1B). Well-watered plantlets under normal growth conditions serve as controls.
- After 14 days of water-withhold treatment, take one or two young leaves for biochemical parameter analysis.
Note: For crude protein/enzyme extraction, the leaves are collected from the same position (second to third leaf from the top) to avoid initial variance among different lines. - Crude protein/enzyme extract
- Weigh 0.2 g of fresh leaf with primary veins from one or two young leaves, grind them with a mortar and pestle in liquid nitrogen (Figure 2A).
Note: Usually, the two youngest leaves collected are enough, and the extra are discarded. Young leaf with a size of 3.5 x 3.5 cm is about 0.1-0.12 g, while a leaf with 4.0 x 4.0 cm is about 0.16-0.19 g. - Homogenize the leaf powder by adding 3 ml of 100 mM PBS buffer (pH 7.8) (Figure 2B).
Note: Unless otherwise specified, all solutions except for NaH2PO4 and Na2HPO4 stock solutions in this protocol are prepared fresh on the day and stored at room temperature during the assay. To simplify the protocol here, use PBS instead of 3% sulphosalicylic acid and 0.25% thiobarbituric acid made in 10% trichloroacetic acid to extract proline and malondialdehyde, respectively. PBS buffer will be frozen after adding the liquid nitrogen. Homogenize the sample in PBS buffer when it begins to thaw in 3-5 min. - Transfer the homogenate to two 1.5 ml centrifuge tubes (Figure 2C) and centrifuge at 10,000 x g for 20 min at 4 °C.
Note: During crude protein/enzyme extraction, make sure all samples are kept under 4 °C or on ice. - Transfer the supernatant (Figure 2D) to new centrifuge tubes for further analysis.
Note: Keep the extract on ice before storing. Aliquot crude protein/enzyme extraction into 1.5 ml centrifuge tubes and drop the tubes in liquid nitrogen to quickly freeze the samples, and then store them at -80 °C. Each aliquot is only used once with one cycle of thawing. - Measure the concentration of crude protein (mg/ml) in the supernatant with Analytik Jena ScanDrop 250 by a spectrophotometric method with Formula Warburg-Christian (protein): protein concentration (mg/ml) = 1.55 x A280 - 0.76 x A260 (Simonian and Smith, 2006) (Figure 3).
Note: Because stray light can affect the linearity of absorbance versus concentration, samples with absorbance > 2.0 should be diluted further in the PBS buffer to obtain absorbance < 2.0 (Simonian and Smith, 2006). - Determination of proline content
- Reagents preparation (see Recipes)
100 mM PBS (pH 7.0)
2.5% acid-ninhydrin
3% sulphosalicylic acid - Prepare reaction solution (for 40 reactions) which contains 10 ml 3% sulphosalicylic, 10 ml acetic acid and 20 ml 2.5% acid-ninhydrin.
- Add 50 μl crude protein/enzyme extract from each sample into 1 ml reaction solution in a 1.5 ml centrifuge tube. Reaction solution with 50 μl 100 mM PBS (pH 7.8) serves as a reference.
- Boil the reaction mixture in a boiling water bath for 15 min.
Note: The reaction mixture turns red after boiling. - Cool down the reaction mixture on ice for 5 min.
- Pipette 200 μl of the reaction mixture and measure the absorbance at 520 nm with Analytik Jena ScanDrop 250.
- Prepare L-proline standard curve for quantification according to the above steps 8b-f (Figure 4).
- Determine proline content of testing sample against the standard curve of L-proline.
- Normalize proline content to protein content in sample with the unit “μg/mg protein”.
- Figure 5A shows the proline content decreased in GbMYB5-silenced cotton under drought stress.
- Determination of malondialdehyde (MDA) content
- Reagents preparation (see Recipes)
10% TCA
0.25% TBA - Add 100 μl crude protein/enzyme extract from each sample into 1 ml 0.25% TBA solution in a 1.5 ml centrifuge tube. A total of 1 ml 0.25% TBA solution with 100 μl 100 mM PBS (pH 7.8) serves as a reference.
- Boil the reaction mixture in a boiling water bath for 15 min.
Note: The reaction mixture turns red after boiling. - Cool down reaction mixture on ice for 5 min.
- Pipette 200 μl of the reaction mixture and measure the absorbance at 532 nm and 600 nm with Analytik Jena ScanDrop 250.
- Formula: MDA (nmol/mg protein) = (A532-A600) x Vr x (V/Vt)/155 x 1,000/(Cp × V)
A532: the absorbance at 532 nm
A600: the absorbance at 600 nm
Vr: the volume of reaction mixture
V: total volume of crude enzyme solution
Vt: volume of crude enzyme used in the testing tube
Cp: crude protein concentration (mg/ml)
155: the extinction coefficient of MDA-TBA at 532 (mM-1 cm-1) - Figure 5B shows the MDA content increased in GbMYB5- silenced cotton under drought stress.
- SOD activity test
- Reagents preparation (see Recipes)
100 mM PBS (pH 7.8)
1 mM EDTA-2Na
130 mM methionine
750 μM NBT
20 μM riboflavin - Prepare reaction solution (for 30 reactions) which contains 30 ml 100 mM PBS (pH 7.8), 0.6 ml 1 mM EDTA-2Na, 2 ml 130 mM Met, 2 ml 750 μM NBT, and 2 ml 20 μM Riboflavin.
- Add 50 μl crude enzyme solution from each sample into 1 ml reaction solution in a 1.5 ml centrifuge tube. Reaction solution with 50 μl 100 mM PBS (pH 7.8) but no crude enzyme under dark and light condition serve as controls I and control II, respectively.
- Put the tubes as well as Control II under light, uniformly, with the light intensity of 4,000 lux for 10-15 min. By contrast, keep Control I in the dark.
- Move the tubes away from the light quickly.
- Measure the absorbance at 560 nm in the dark with Analytik Jena ScanDrop 250. Use Control I as reference.
- Formula: SOD total activity (unit: u/mg protein) = [(Ack-As) x V]/(0.5 x Ack x Vt)/(Cp × V)
Ack: the absorbance at 560 nm of Control II (exposed to light with no crude enzyme)
As: the absorbance at 560 nm of candidate sample tube
V: total volume of crude enzyme solution
Vt: volume of crude enzyme used in the testing tube
Cp: crude protein concentration (mg/ml)
0.5: One unit of SOD is defined as the amount of enzyme that inhibits 50% nitroblue tetrazolium photoreduction. - Figure 5C shows the activities of SOD decreased in GbMYB5-silenced cotton under drought stress.
- POD activity test
- Reagents preparation (see Recipes)
100 mM PBS (pH 7.0)
0.2% guaiacol
30% H2O2 - Prepare reaction solution (for 50 reactions) by adding 28 μl 0.2% guaiacol in 50 ml 100 mM PBS (pH 7.0), heat and stir well, add 19 μl 30% H2O2 after cooling.
- Add 50 μl 100 mM PBS (pH 7.8) and 1 ml of the reaction solution into a cuvette for reference (see the operation video).
Note: Use a pipettor and tip to remove the solutions from the cuvette before measuring the next sample.Video 1. Enzyme activity determination by spectrophotometer ScanDrop 250 (Take POD for example). This smart phone video provides a whole procedure of enzyme activity determination by spectrophotometer ScanDrop 250: how to organize the working batch, how to store all reaction solutions and crude extracts, how to set the measurement parameters in the software, how to add testing crude extracts and reaction buffer, etc. The enzyme activity determination of CAT and GST is the same as the POD.
Note: In order to make a homogeneous solution for enzyme reaction, it is importance to add the crude extracts (small volume) in prior to the reaction buffer (big volume) into the cuvette. - Add 50 μl crude enzyme solution into a cuvette, insert the cuvette into the holder, then add 1 ml of the reaction solution into the cuvette and immediately record the dynamic absorbance at 470 nm with Analytik Jena ScanDrop 250 at every 15 sec for 1 min, looking for steady average alteration.
Note: The best linear regression between absorbance at 470 nm and reaction time is within 75 sec (Figure 6). - Formula: POD activity (unit: u/mg protein) = △A470 x (V/Vt)/(0.01 x t)/(Cp × V)
△A470: the change of absorbance at 470 nm during every 15 sec
V: total volume of crude enzyme solution
Vt: volume of crude enzyme used in the testing tube
t: reaction time (min)
Cp: crude protein concentration (mg/ml)
0.01: One unit of POD is defined as the amount of enzyme that increases 0.01 of absorbance at 470 nm per minute. - Figure 5D shows the activities of POD decreased in GbMYB5-silenced cotton under drought stress.
- CAT activity assay
- Reagents preparation
100 mM PBS (pH 7.0)
30% H2O2 - Prepare reaction solution (for 50 reactions) by adding 77.5 μl 30% H2O2 in 50 ml 100 mM PBS (pH 7.0).
- Add 50 μl crude enzyme into a cuvette, insert the cuvette into the holder, then add 1 ml of the reaction solution into the cuvette and immediately record the dynamic absorbance at 240 nm with Analytik Jena ScanDrop 250 at every 15 sec for 1 min, looking for steady average alteration. Reaction solution with 50 μl 100 mM PBS (pH 7.8) serves as a reference.
- Formula: CAT activity (unit: u/mg protein) = △A240 x (V/Vt)/(0.1 x t)/(Cp × V)
△A240: the change of absorbance at 240 nm during every 15 sec
V: total volume of crude enzyme solution
Vt: volume of crude enzyme used in the testing tube
t: reaction time (min)
Cp: crude protein concentration (mg/ml)
0.1: One unit of CAT is defined as the amount of enzyme that decreases 0.1 of absorbance at 240 nm per minute. - Figure 5E shows the activities of CAT decreased in GbMYB5-silenced cotton under drought stress.
- GST activity assay
- Reagents preparation (see Recipes)
100 mM PBS (pH 6.5)
5 mM GSH
1.5 mM CDNB - Prepare reaction solution (for 50 reactions) by adding 20 ml 5 mM GSH and 40 ml 1.5 mM CDNB.
- Add 50 μl crude enzyme solution into a cuvette, insert the cuvette into the holder, then add 1 ml of the reaction solution into the cuvette and immediately record the dynamic absorbance at 340 nm with Analytik Jena ScanDrop 250 at every 15 sec for 1 min, looking for steady average alteration. Reaction solution with 50 μl 100 mM PBS (pH 7.8) serves as a control and ddH2O serves as a reference.
- Formula: GST activity (unit: u/mg protein) = (△A340s - △A340ck) x (V/Vt)/t/(Cp × V)
△A340s: the change of A340 of candidate sample tube during every 15 sec
△A340ck: the change of A340 of control during every 15 sec
V: total volume of crude enzyme solution
Vt: volume of crude enzyme used in the testing tube
t: reaction time (min)
Cp: crude protein concentration (mg/ml)
One unit of GST is defined as the amount of enzyme that increases 1 of absorbance at 340 nm per min. - Figure 5F shows the activities of GST decreased in GbMYB5-silenced cotton under drought stress.
Representative data
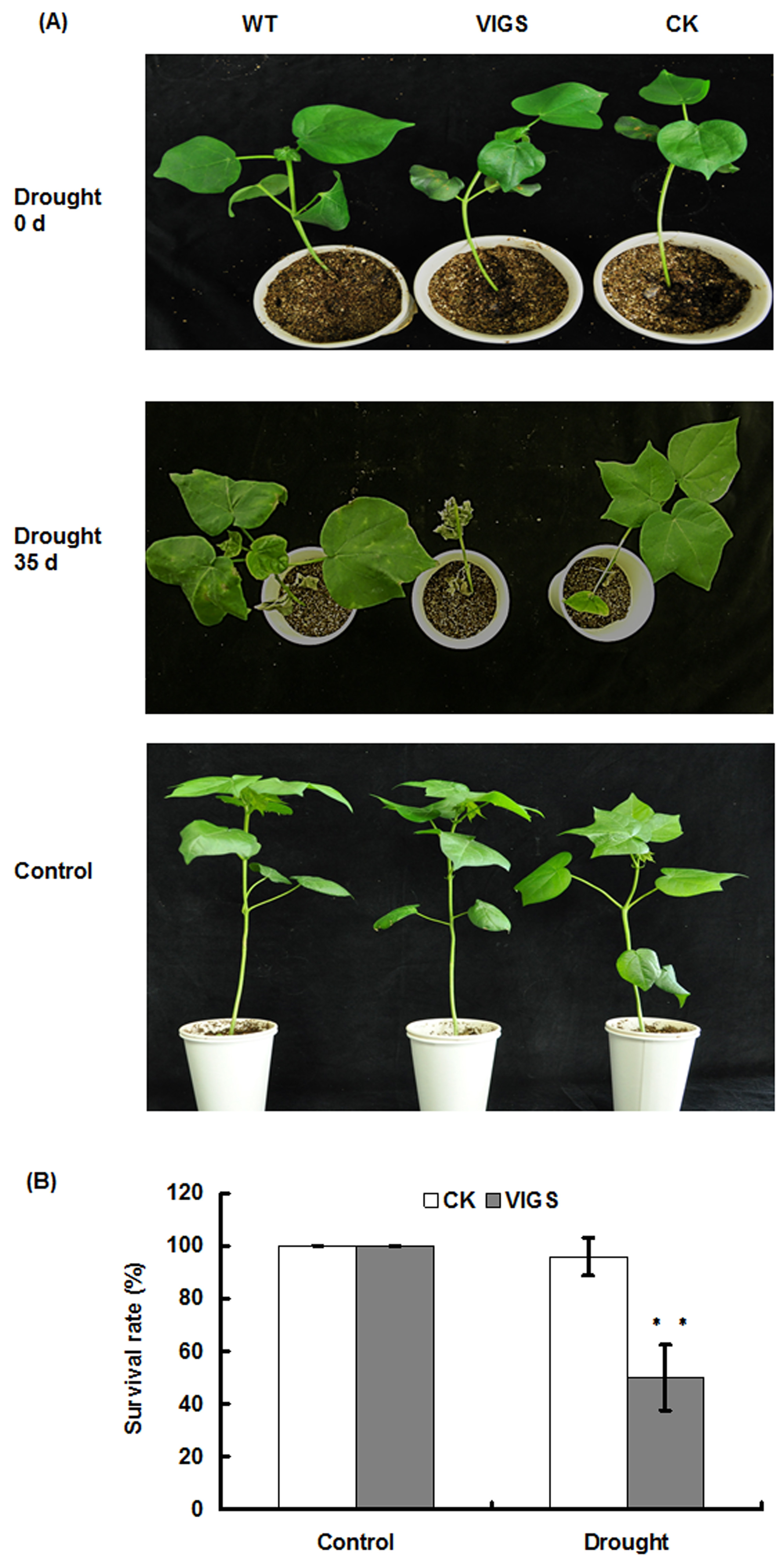
Figure 1. Phenotypes (A) and survival rates (B) of GbMYB5-silenced cotton plantlets under water-withholding treatment. A. The phenotype of cotton plantlets in response to water-withhold treatment. WT: wild-type cotton plantlets; VIGS: cotton plantlets agroinfiltrated with the vectors pCLCrVA-GbMYB5 and pCLCrVB to silence the GbMYB5 gene (Chen et al., 2015); CK: cotton plantlets agroinfiltrated with the empty vectors pCLCrVA and pCLCrVB. 0 d: the beginning of water-withholding treatment; 35 d: 35 d post water-withholding treatment; Control: normal growth conditions. B. The survival rates of cotton plantlets after 35 d of water-withholding treatment. Control: normal growth conditions; Drought: 35 d post water-withholding treatment; The survival values are presented as means ± SE from three biological replicates with eight plants per replicate (**P < 0.01; t-test).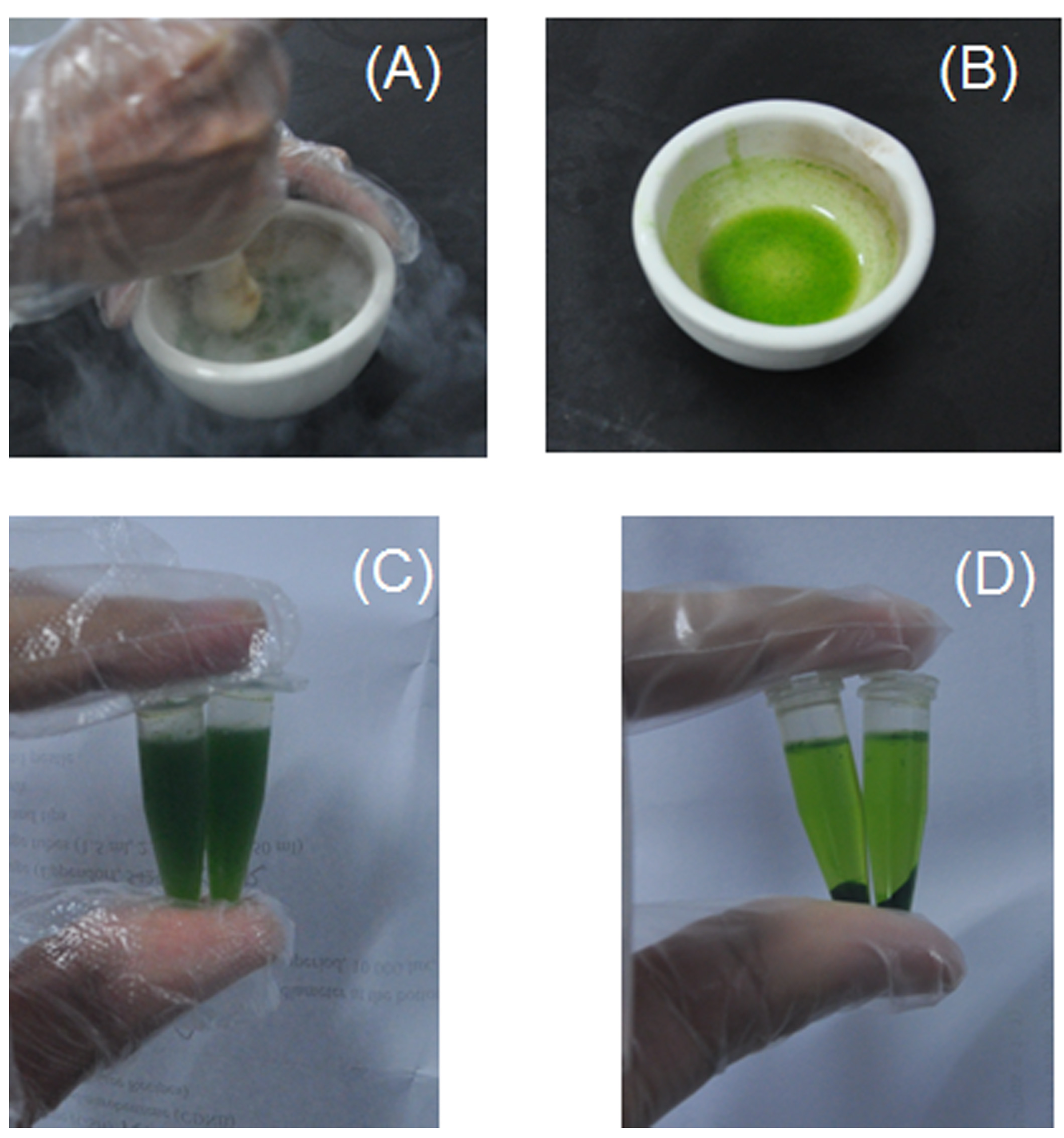
Figure 2. Crude protein/enzyme extraction from cotton leaves of 14 d post water-withholding treatment. A. Grind leaf tissue with a mortar and pestle in liquid nitrogen. B. Homogenize the leaf powder with PBS buffer. C. Tissue homogenates in centrifuge tubes before centrifugation. D. Supernatants and pellets after centrifugation.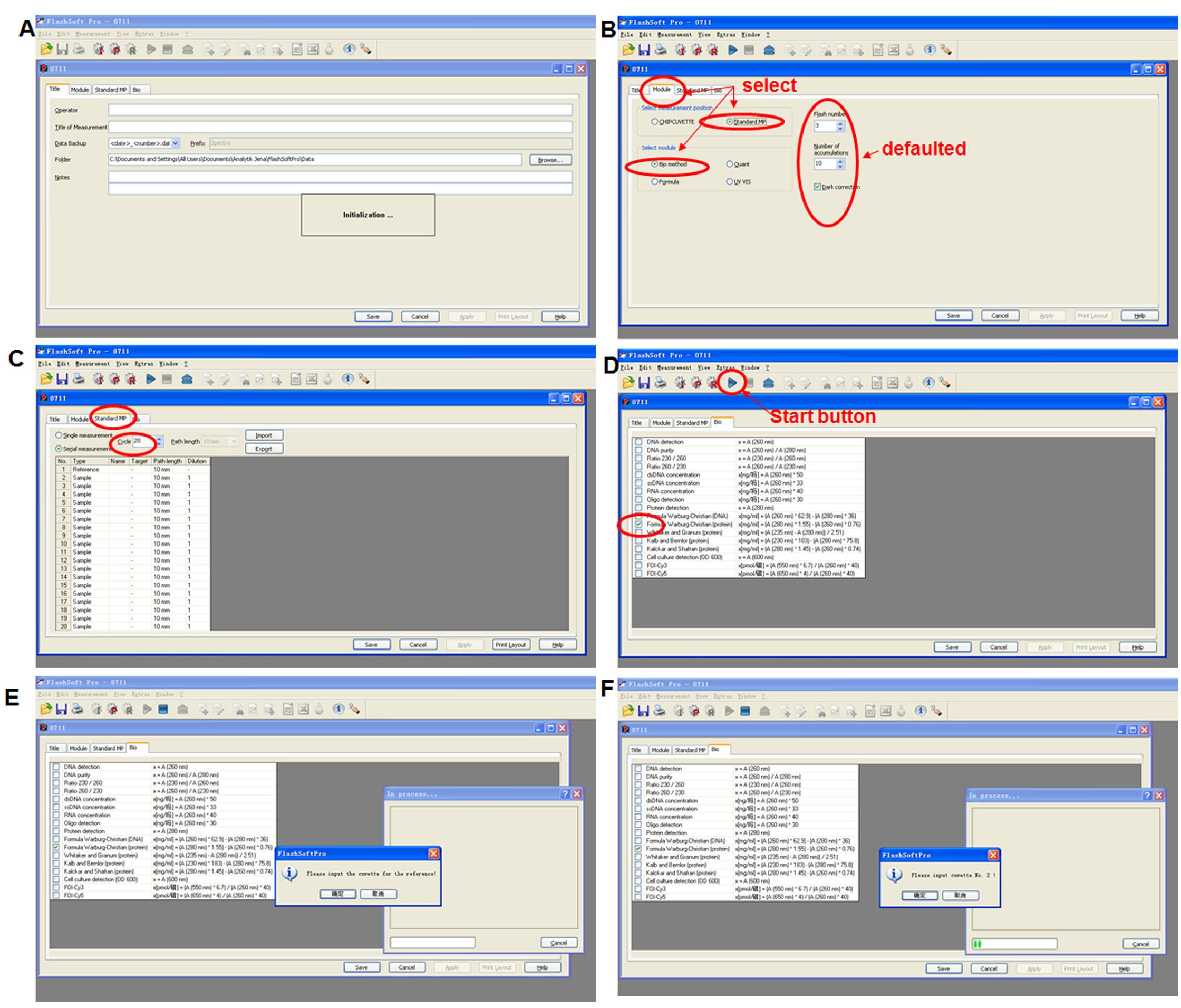
Figure 3. Protein quantification with spectrophotometer ScanDrop 250. A. Start machine ScanDrop 250 and its software FlashSoft Pro. B. Select “Standard MP” measurement position and “Bio method” module in the Module window, set “Flash number” (3),“Number of accumulation” (10) and activate “Dark correction”. C. Select the “Serial measurement”, enter the number of samples under “Cycle” in the Standard MP window. D. Activate the desired Formula Warburg-Christian (protein) by ticking the box in the “Bio” window, then press start button. E. Insert cuvette for reference (100 mM PBS buffer, pH = 7.8) according to the instruction. F. Measure the absorbance of the crude solutions according to the instruction. The key points of operation are marked with red circles.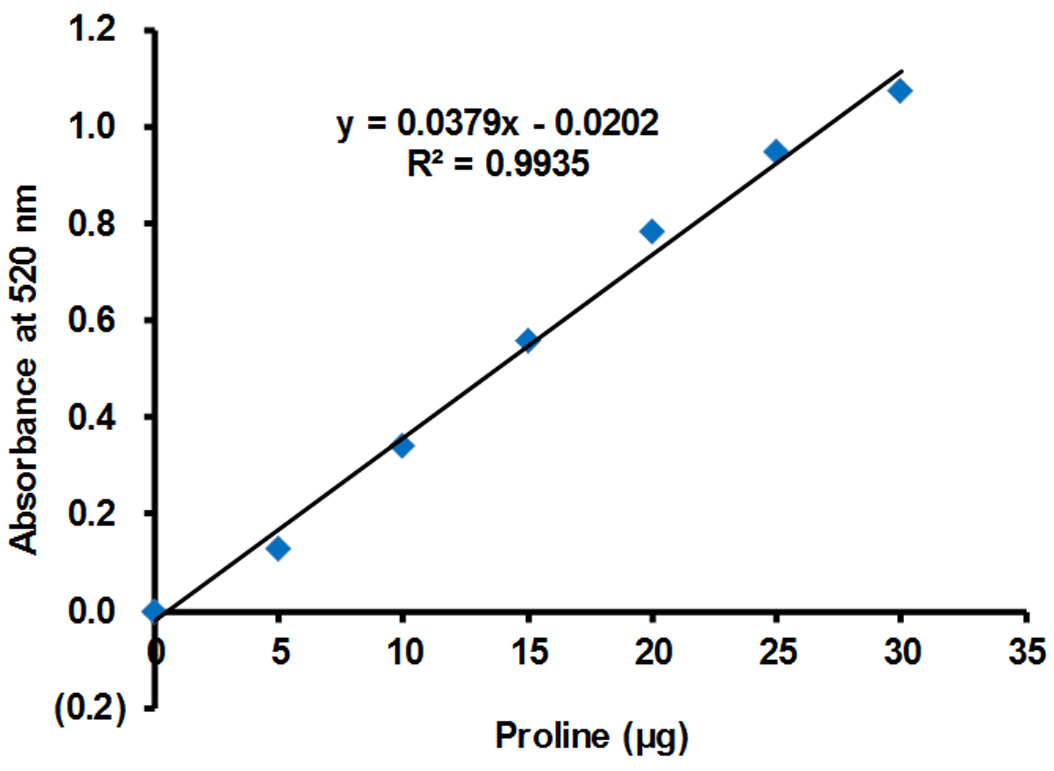
Figure 4. The standard curve between the absorbance at 520 nm and L-proline content. Series content of L-proline (0, 5, 10, 15, 20, 25, and 30 μg) dissolved in 0.5 ml ddH2O were added into 1 ml of reaction solution to measure their absorbance at 520 nm. A linear regression was observed between the absorbance values at 520 nm and L-proline contents at 0-30 μg (R2 = 0.9935). The values are means of two independent experiments.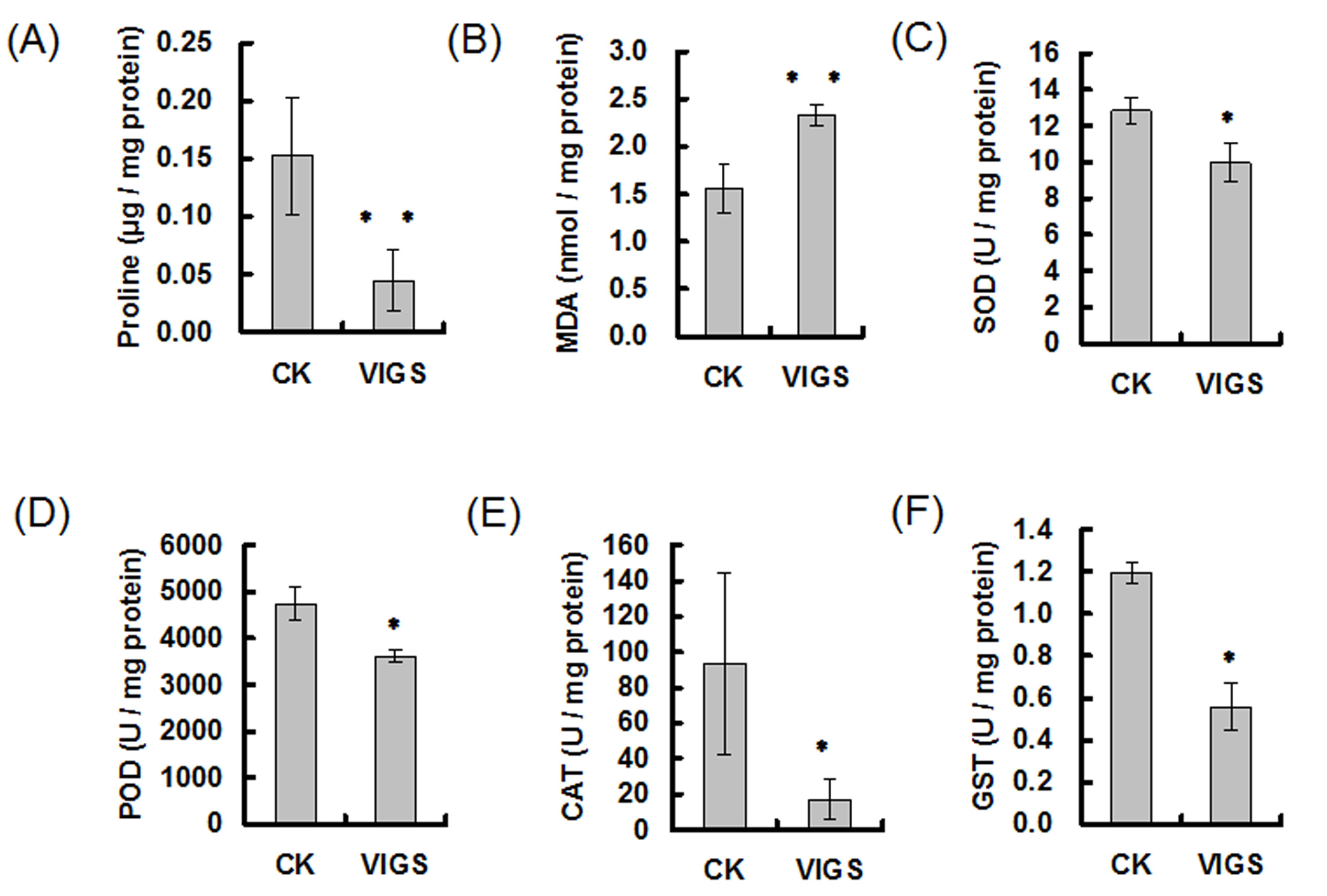
Figure 5. Analysis of metabolites and enzyme activities in GbMYB5-silenced cotton under drought stress. A. Proline content decreased in GbMYB5-silenced cotton under drought stress. B. MDA content increased in GbMYB5- silenced cotton under drought stress. Activities of SOD (C), POD (D), CAT (E), and GST (F) decreased in GbMYB5-silenced cotton under drought stress. CK: cotton plantlets agroinfiltrated with the empty vectors pCLCrVA and pCLCrVB; VIGS: cotton plantlets agroinfiltrated with the vectors pCLCrVA-GbMYB5 and pCLCrVB to silence the GbMYB5 gene. Values were means ± SE of biological replicates (n ≥ 5) (*P < 0.05; **P < 0.01; t-test).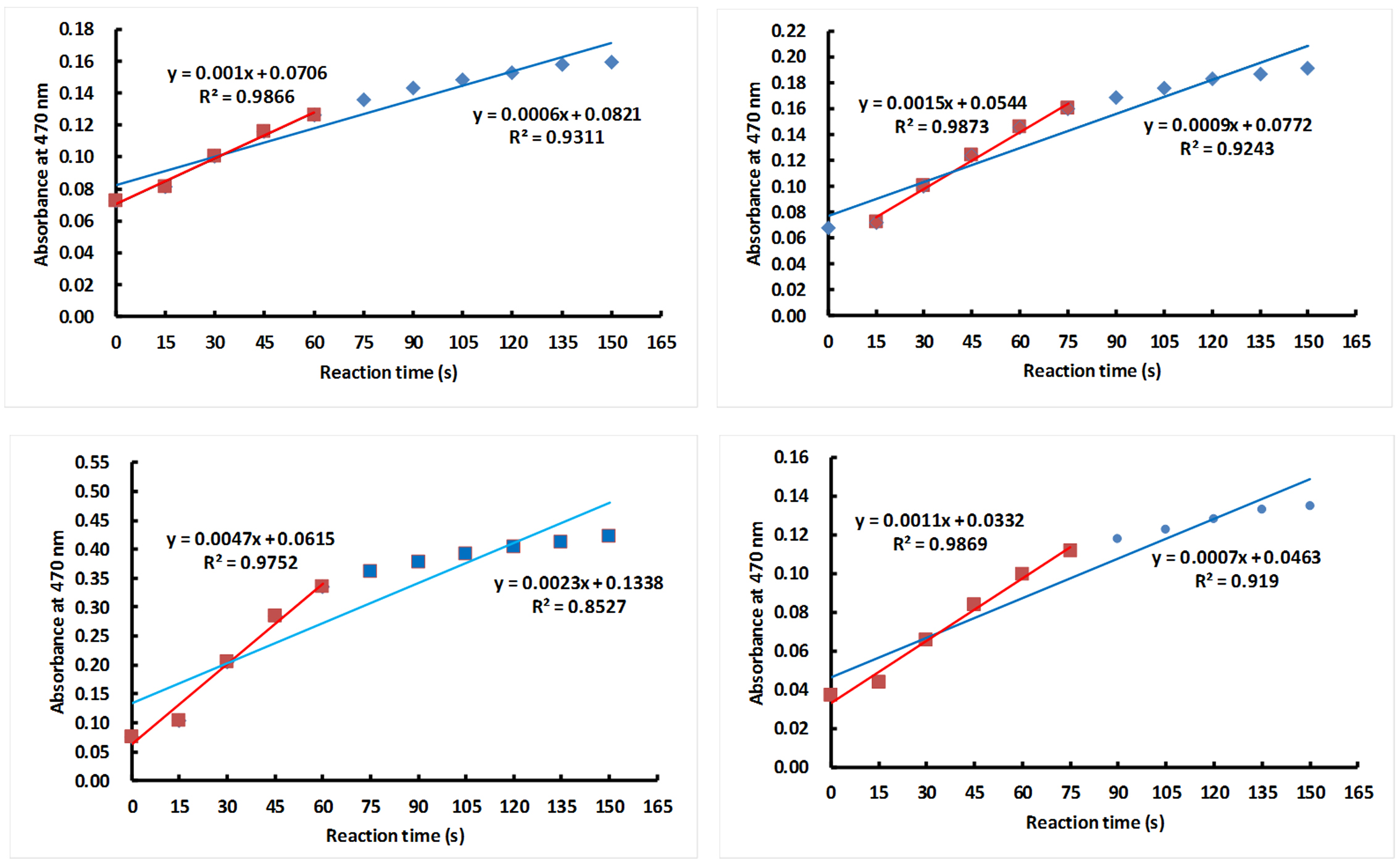
Figure 6. Linear regression between absorbance at 470 nm and reaction time. The absorbance value of POD was automatically collected in a 15 sec interval and up to 150 sec. The blue line and red line represent the linear regression between absorbance of POD at 470 nm and reaction time within 150 sec or 60-75 sec, respectively. The tests of 4 POD samples (4 sub-panels) shows the best linear regression between absorbance at 470 nm and reaction time is within 75 sec.
Recipes
Note: The pH value of the PBS buffer should be verified with pH meter before using.
- 1/2 MS solution
Dissolve 2.47 g 1/2 MS media salt in 1,000 ml ddH2O - 200 mM Na2HPO4 stock solution
Dissolve 53.65 g Na2HPO4·7H2O in 1,000 ml ddH2O
Keep in 4 °C before using - 200 mM NaH2PO4 stock solution
Dissolve 27.8 g NaH2PO4·H2O in 1,000 ml ddH2O
Keep in 4 °C before using - 100 mM PBS (pH 7.0)
61 ml 200 mM Na2HPO4 stock solution
39 ml 200 mM NaH2PO4 stock solution
100 ml ddH2O - 2.5% acid-ninhydrin
Dissolve 1.250 g ninhydrin in 30 ml acetic acid and 20 ml 100 mM PBS (pH 7.0) stirring in 70 °C water bath
Keep in 4 °C before using - 3% sulphosalicylic acid
Dissolve 3.496 g 5-sulphosalicylic acid dihydrate in 100 ml ddH2O - 10% TCA
Dissolve 10 g TCA in 100 ml ddH2O - 0.25% TBA
Dissolve 0.125 g TBA in 5 ml 1 mol/L NaOH
Add into 45 ml 10% TCA
Keep in 4 °C before using - 100 mM PBS (pH 7.8)
91.5 ml 200 mM Na2HPO4 stock solution
8.5 ml 200 mM NaH2PO4 stock solution
100 ml ddH2O - 1 mM EDTA-2Na
Dissolve 0.037 g EDTA-2Na in 100 ml ddH2O - 130 mM methionine
Dissolve 0.970 g methionine in 50 ml 100 mM PBS (pH 7.8)
Keep in 4 °C before using - 750 μM NBT
Dissolve 0.031 g NBT in 50 ml 100 mM PBS (pH 7.8)
Keep in the dark - 20 μM Riboflavin
Dissolve 0.007 g riboflabin in 100 ml ddH2O
Keep in the dark - 0.2% guaiacol
Dissolve 0.1 g guaiacol in 0.5 ml ethanol
Add into 50 ml 100 mM PBS (pH 7.0)
Keep in the dark - 100 mM PBS (pH 6.5)
31.5 ml 200 mM Na2HPO4 stock solution
68.5 ml 200 mM NaH2PO4 stock solution
100 ml ddH2O - 5 mM GSH
Dissolve 0.077 g GSH in 50 ml 100 mM PBS (pH 6.5) - 1.5 mM CDNB
Dissolve 0.015 g CDNB in 5 ml ethanol
Add into 45 ml 100 mM PBS (pH 6.5)
Acknowledgments
This protocol was adapted from the research article: Chen et al. (2015). This work was supported by the National Natural Science Foundation of China [grant No.31371930]; National Science and Technology Major Project for Transgenic Breeding, China [grant No.2014ZX0800501B]; the Independent Innovation of Agricultural Sciences in Jiangsu Province, China [grant No.CX(15)1005].
References
- Chen, T., Li, W., Hu, X., Guo, J., Liu, A. and Zhang, B. (2015). A cotton MYB transcription factor, GbMYB5, is positively involved in plant adaptive response to drought stress. Plant Cell Physiol 56(5): 917-929.
- Gao, X., Wheeler, T., Li, Z., Kenerley, C. M., He, P. and Shan, L. (2011). Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J 66(2): 293-305.
- Gu, Z., Huang, C., Li, F. and Zhou, X. (2014). A versatile system for functional analysis of genes and microRNAs in cotton. Plant Biotechnol J 12(5): 638-649.
- Simonian, M. H. and Smith, J. A. (2006). Spectrophotometric and colorimetric determination of protein concentration. Curr Protoc Mol Biol Chapter 10: Unit 10 11A.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Chen, T. and Zhang, B. (2016). Measurements of Proline and Malondialdehyde Content and Antioxidant Enzyme Activities in Leaves of Drought Stressed Cotton. Bio-protocol 6(17): e1913. DOI: 10.21769/BioProtoc.1913.
Category
Plant Science > Plant physiology > Abiotic stress
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link