- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Determination of the H+-ATP Synthase and Hydrolytic Activities
(*contributed equally to this work) Published: Vol 6, Iss 16, Aug 20, 2016 DOI: 10.21769/BioProtoc.1905 Views: 10311
Reviewed by: Masahiro MoritaDaniel KrausAntoine de Morree

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Measurement of the Activity of Wildtype and Disease-Causing ALPK1 Mutants in Transfected Cells With a 96-Well Format NF-κB/AP-1 Reporter Assay
Tom Snelling
Nov 20, 2024 1611 Views

Fluorescence Polarization-Based High-Throughput Screening Assay for Inhibitors Targeting Cathepsin L
Keyu Guo [...] Shuyi Si
Jul 20, 2025 2282 Views

Detecting the Activation of Endogenous Small GTPases via Fluorescent Signals Utilizing a Split mNeonGreen: Small GTPase ActIvitY ANalyzing (SAIYAN) System
Miharu Maeda and Kota Saito
Jan 5, 2026 496 Views
Abstract
The H+-ATP synthase of the inner mitochondrial membrane utilizes the proton gradient generated by the respiratory chain to synthesize ATP. Under depolarizing conditions, it can function in reverse by hydrolyzing ATP to generate a proton gradient. The protocols presented here allow the facile determination of both the synthetic and hydrolytic activities of the H+-ATP synthase in isolated mitochondria and in permeabilized mammalian cells. Since the protocol requires the isolation of polarized and well-coupled mitochondria, first we describe the protocol for mitochondrial isolation from mouse tissues. Second, we describe the protocol for measuring the ATP synthetic activity as end-point and kinetic modes in isolated mitochondria and in permeabilized cells. Finally, we describe the protocol for the determination of the ATP hydrolytic activity of the enzyme in isolated mitochondria.
Keywords: MitochondriaPart I. The isolation of mitochondria from mouse tissues
Materials and Reagents
- 10 ml tubes
- 1.5 ml Eppendorf tubes
- Sucrose (Merck Millipore Corporation, catalog number: 107651 )
- Ethylenediaminetetraacetic acid disodium salt dehydrate (Sigma-Aldrich, catalog number: ED2SS )
- D-Sorbitol (Sigma-Aldrich, catalog number: S1876 )
- Ethylene glycol-bis (2-aminoethylether)-N,N,N′,N′-tetraacetic acid (Sigma-Aldrich, catalog number: E4378 )
- Trizma® base (Sigma-Aldrich, catalog number: T1503 )
- Bovine serum albumin (Sigma-Aldrich, catalog number: A7906 )
- Protein assay dye reagent concentrate (Bio-Rad Laboratories, catalog number: 5000006 )
- Medium A (see Recipes)
- Medium H (see Recipes)
Equipment
- 15 ml Dounce All-Glass tissue grinder (Kimble, catalog number: 885303-0015 )
- Beckman Avanti J-25 centrifuge (Beckman Coulter, model: Avanti J-25 )
- JA-25.50 rotor, fixed angle, aluminum, single lock lid, 8 x 50 ml, 25,000 rpm, 75,600 x g (Beckman Coulter, catalog number: 363055 )
- Bio-VialTM tube, PP, 4 ml, 14 x 55 mm, 0.56 x 2.2 in (Beckman Coulter, catalog number: 566353 )
- Centrifuge 5415R (Sigma-Aldrich, Eppendorf®, model: 5415R )
Procedure
- Animal studies were carried out in compliance with animal policies and ethical guidelines of the European Community. The project was approved by the Institutional Review Board (Ethical Committee of the UAM, CEI-24-571).
- The isolation of mitochondria from mouse liver and/or heart was performed according to (Fernandez-Vizarra et al., 2010) with minor modifications.
- Adult mice are sacrificed by cervical dislocation and the liver (roughly half of the liver without the gallbladder) or the whole heart is removed and immediately cooled down at 4 °C in the homogenization medium A (liver) or medium H (heart). At this point, place several 10 ml centrifuge tubes on ice until step 6.
- The tissue is washed, weighted and minced into small pieces with a pair of scissors in precooled 1x PBS to remove blood and connective tissue.
- The homogenization is performed in the tissue grinder with 4 ml/g of cold homogenization medium A (liver) or with 10 ml/g of cold homogenization medium H (heart). It is necessary to make 5 steps with pestle A followed by 5 steps with pestle B in the case of liver and 12 steps with pestle A followed by 12 steps with pestle B in the case of heart.
- The homogenate is transferred to the precooled 10 ml tubes and centrifuged for 10 min at 1,000 x g at 4 °C. This step is then repeated with the supernatant in order to discard unbroken tissue, cells and nuclei.
- The resulting supernatant is transferred to 1.5 ml Eppendorf tubes and centrifuged in a microfuge for 10 min at 6,700 x g at 4 °C to pellet the mitochondria.
Notes
- It is very important to perform the entire isolation process at 4 °C using cold medium and without long intermediate stops.
- Mitochondria should not be centrifuged any more in order to preserve functional integrity of the organelle.
Recipes
- Medium A (100 ml)
0.32 M sucrose (MW = 342.2965 g/mol): 10.95 g
1 mM EDTA (MW = 292.24 g/mol): 30 mg
10 mM Tris-HCl (pH 7.4) (MW = 121.14 g/mol): 0.12 g - Medium H (100 ml)
0.072 M sucrose (MW = 342.2965 g/mol): 2.46 g
0.225 M sorbitol (MW = 182.17 g/mol): 4.1 g
1 mM EGTA (MW = 380.4 g/mol): 40 mg
0.1% w/v fatty acid-free bovine serum albumin (BSA): 0.1 g
10 mM Tris-HCl (pH 7.4) (MW = 121.14 g/mol): 0.12 g
Materials and Reagents
- Microplate, 96 well, Polystyrene, black flat bottom, TC-treated (Cultek, catalog number: 153603 )
- Protein assay dye reagent concentrate (Bio-Rad Laboratories, catalog number: 5000006 )
- Phosphatase inhibitor cocktail 2 (Sigma-Aldrich, catalog number: P5726 )
- cOmpleteTM, Mini, EDTA-free (Sigma-Aldrich, Roche Diagnostics®, catalog number: 11836170001 )
- Digitonin (Sigma-Aldrich, catalog number: D141 )
- P1, P5-di(adenosine-5′) pentaphosphate pentasodium salt (Sigma-Aldrich, catalog number: D4022 )
- Adenosine 5′-diphosphate sodium salt (Sigma-Aldrich, catalog number: A2754 )
- Adenosine 5′-triphosphate (ATP) disodium salt hydrate (Sigma-Aldrich, catalog number: FLAAS )
- HClO4
- KOH
- Oligomycin (VWR International, catalog number: 80058-538 )
- Succinic acid (Sigma-Aldrich, catalog number: 398055 )
- Rotenone (Sigma-Aldrich, catalog number: R8875 )
- ATP bioluminescence assay kit CLS II (Roche Diagnostics, catalog number: 11699695001 )
- Luciferase from Photinus pyralis (Sigma-Aldrich, catalog number: 10411523001 )
- Beetle luciferin, potassium salt (Promega, catalog number: E1601 )
- pH indicator solution (pH 4.0-10.0) (Merck Millipore Corporation, catalog number: 1091750100 )
- Trizma® base (Sigma-Aldrich, catalog number: T1503 )
- Bovine serum albumin (Sigma-Aldrich, catalog number: A7906 )
- Sucrose (Merck Millipore Corporation, catalog number: 107651 )
- KCl (Merck Millipore Corporation, catalog number: 104936 )
- MgCl2 (Merck Millipore Corporation, catalog number: 172571 )
- Phosphate-potassium buffer (Sigma-Aldrich, catalog number: P0662 / P3786 )
- Ethylene glycol-bis (2-aminoethylether)-N,N,N′,N′-tetraacetic acid (EGTA) (Sigma Aldrich, catalog number: E4378 )
- Respiration Medium (RM) (see Recipes)
Equipment
- FLUOstar Omega Microplate Reader (BMG Labtech, model: FLUOstar Omega )
- Heraeus Megafuge 11R (Thermo Fisher Scientific, model: Heraeus Megafuge 11R )
- Centrifuge 5415R (Sigma-Aldrich, Eppendorf®, model: 5415R )
Procedure
- End Point determination of the ATP synthetic activity
- If working with isolated mitochondria from animal tissues the isolated organelles are resuspended at 1 µg/µl of protein in Respiration Medium (RM) containing 1x phosphatase and protease inhibitors. The mitochondrial preparation is kept at 4 °C. No further procedures are required until step A7.
- If working with cells, the cells (~1-3 x 106 cells) are trypsinized and washed twice with PBS and the cellular pellet resuspended in 1 ml of RM at RT containing 1x phosphatase and protease inhibitors. Cellular permeabilization is achieved by addition of digitonin at a concentration of 50-75 µg/ml of RM.
- Tubes are shaken by repeated inversions during 1 min. For the removal of digitonin, immediate centrifugation of the cells at 100 x g during 5 min followed by two washes with RM at room temperature is required.
- At this point, a small fraction of the permeabilized cells are separated and used for the quantification of total protein concentration using a colorimetric based assay such as Bradford Protein Assay or the Bicinchoninic acid Assay (BCA).
- Prepare two sets of tubes; one is needed for the reaction without oligomycin (OL) and one for the parallel reaction with oligomycin. In addition, prepare two sets of seven tubes each containing 200 μl of 6% HClO4 to be used under step A9.
- RM is used for the preparation of the reaction buffer by supplementation with 150 µM P1, P5-di(adenosine-5′) pentaphosphate (an adenylate kinase inhibitor that prevents conversion of ADP into ATP), 2 mM rotenone, 5 mM ADP and 2 mM succinate as respiratory substrate (Complex II). Other respiratory substrates can be used.
- The parallel reaction using reaction buffer supplemented with 30 µM OL is assayed for each sample. This reaction indicates the ATP synthesized that might occur independent of the H+-ATP synthase activity.
- Triggering of the reaction of ATP synthesis is achieved by the addition of 150-200 μg of protein of permeabilized cells or of 50 μg of the isolated mitochondria to the tube containing 450 μl of the reaction buffer (as described under steps A6 and A7) in a tube shaker at constant shaking and at 30 °C.
- Small aliquots (50 μl) are taken every 30 sec until 3 min (end of reaction) and added to the tubes containing 200 μl of 6% HClO4.
- The tubes are immediately vortexed and placed at 4 °C during 1 h for the precipitation of proteins.
- Afterwards, tubes are centrifuged at 11,000 x g during 5 min and the resulting supernatant transferred to another tube for neutralization with 10% KOH (~130 µl) in the presence of 2 µl of pH indicator.
- The content of ATP in each sample is determined using the ATP bioluminescence assay kit CLS II in 96-well plates with a luminometer plate-reader. To this aim, mix in each well 50 μl of the neutralized sample with 50 μl of the reaction buffer included in the kit and containing luciferase. An ATP standard curve (0-10 µM) is generated and processed in the same plate adding 50 μl of ATP standard and 50 μl of the reaction buffer included in the kit.
- The initial rate of ATP synthesis is determined by estimation of the pmoles of ATP produced in each sample. The ATP synthetic activity of the enzyme is expressed as nmoles of ATP/min/mg of protein (as indicated in Representative data, Figure 1).
- If working with isolated mitochondria from animal tissues the isolated organelles are resuspended at 1 µg/µl of protein in Respiration Medium (RM) containing 1x phosphatase and protease inhibitors. The mitochondrial preparation is kept at 4 °C. No further procedures are required until step A7.
- Kinetic determination of the ATP synthetic activity
- For assaying the mitochondrial ATP synthetic activity in kinetic mode we followed the protocol detailed in (Vives-Bauza et al., 2007) with some modifications.
- Steps B1-4 were performed according to the previous protocol.
- Permeabilized cells or 50-150 µg of isolated mitochondria are resuspended in 40 µl of RM. The tubes are kept on ice until used.
- Two mixes A and B with the reaction compounds are prepared in RM (see Recipes 2 and 3)
- Add 160 µl of Mix A with 20 µl of Mix B in each well. The luminometer plate-reader is set up to measure luminescence in kinetic mode every 10 sec during ~10 min in which luminescence increases linearly for ~250 sec.
- Place the plate in the luminometer and add 20 µl of permeabilized cells or isolated mitochondria obtained in step B3 to start the reaction of ATP synthesis.
- Relative light units are converted to ATP concentration using an ATP standard curve. To this end, prepare triplicates of ATP solutions between 0 and 10 µM in Mix A at a final volume of 180 µl. Finally, add 20 µl of Mix B to each well and measure luminescence at end point (see Figure 2).
- The initial rate of ATP synthesis is determined by estimation of the pmoles of ATP produced in each sample. The ATP synthetic activity of the enzyme is expressed as pmoles of ATP/min/mg of protein (as indicated in Representative data, Figure 2).
- For assaying the mitochondrial ATP synthetic activity in kinetic mode we followed the protocol detailed in (Vives-Bauza et al., 2007) with some modifications.
Representative data
- To convert the RLU obtained into ATP produced it is necessary to follow the next steps:
- Subtract the RLU in T0, T1, T2 and T3 time points in tubes + OL (a relatively high value because of the ATP contained in mitochondria) from the time points taken in the absence of OL (T0, T1, T2, T3) (Figure 1A). It is very important to verify that RLU increases linearly and significantly in the absence of OL (Figure 1A) whereas subtle or negligible changes in RLU occur in the presence of OL (Figure 1A).
- Interpolate the RLU values obtained (Figure 1A) in the linear regression (Figure 1B) to obtain the ATP content in the volume of sample that has been measured.
- Multiply the pmol of ATP obtained by the fixed value of 7.6, which is the ratio between the total volume and the volume of sample measured. (Total volume = 250 µl + 130 µl = 380 µl); (Volume of sample = 50 µl); (Ratio = 7.6) (from steps A9 and A11).
- Multiply the value obtained under point 3 by the factor that indicates the volume of sample taken at each time point relative to the total volume of reaction left (i.e., Factor 9 at T1 because 450/50 = 9; Factor 8 at T2 because 400/50 = 8; Factor 7 at T3 because 350/50 = 7, ….) (from step A9).
- Normalized the total pmol of ATP obtained by the protein amount and reaction time (Figure 1C).
- The same calculation procedure is followed when the activity of a permeabilized-cell preparation is used.
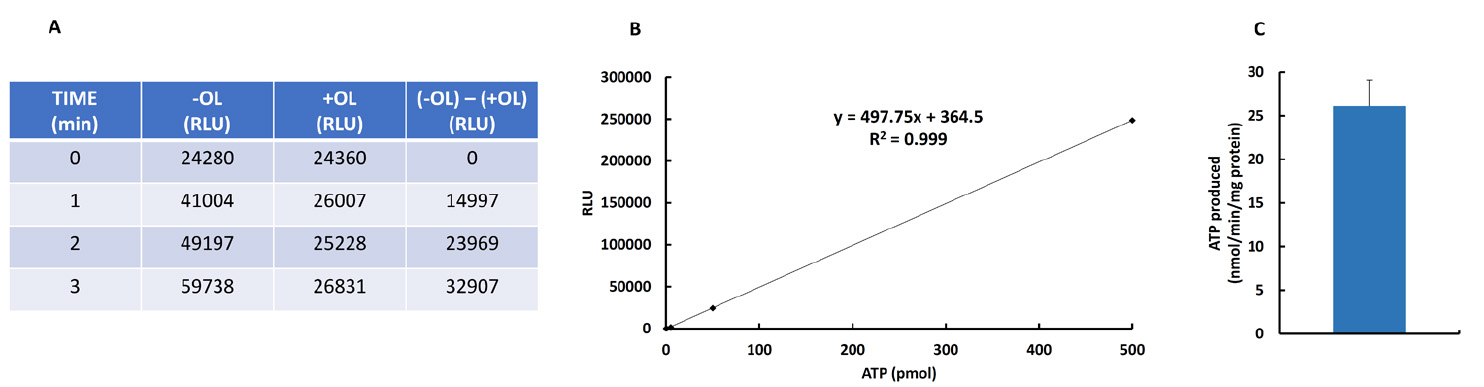
Figure 1. ATP synthase activity (end point mode) in isolated mitochondria from mouse heart in the absence (-OL) or presence (+OL) of oligomycin (OL). A. Relative light units (RLU) of samples at 0, 1, 2 and 3 min after triggering the reaction in the absence and presence of OL. B. Linear regression analysis for the correlation between ATP amount and luciferin/luciferase-dependent luminescence. C. Example of the ATP synthetic activity (mean ± S.E.M) using three preparations of heart mitochondria.
- Subtract the RLU in T0, T1, T2 and T3 time points in tubes + OL (a relatively high value because of the ATP contained in mitochondria) from the time points taken in the absence of OL (T0, T1, T2, T3) (Figure 1A). It is very important to verify that RLU increases linearly and significantly in the absence of OL (Figure 1A) whereas subtle or negligible changes in RLU occur in the presence of OL (Figure 1A).
- To convert the RLU obtained into ATP produced it is necessary to follow the next steps:
- Subtract the RLU at T0 from all other time points (Figure 2A).
- To convert the RLU of the reaction into ATP amount it is necessary to interpolate the RLU at 60 sec in the linear regression (Figure 2B).
- It is important to normalized data according to the protein amount in the sample (Figure 2C).
Note: If you use the RLU at 60 sec to interpolate into Figure 2B, your data is already a rate (pmol ATP/min). - The same calculation procedure is followed when the activity of a preparation of isolated mitochondria is used.
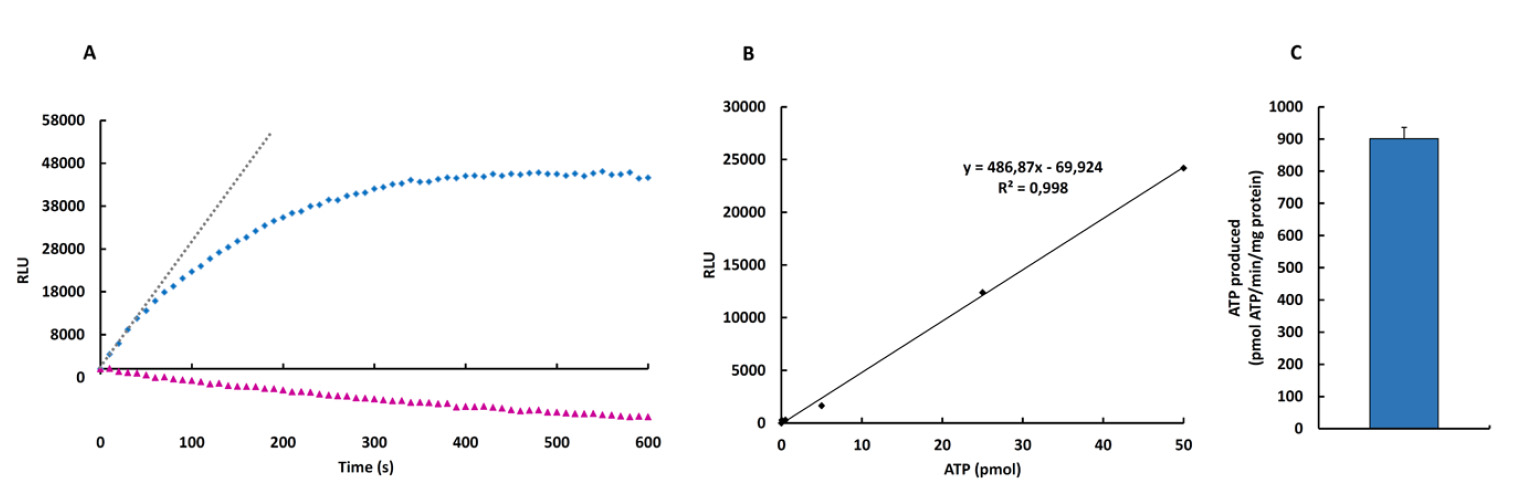
Figure 2. ATP synthase activity (kinetic mode) in digitonin-permeabilized HCT116 cells in the absence (blue) or presence of oligomycin (pink). A. Kinetic representation of the production of ATP in relative light units (RLU). The gray dotted line represents the initial rate of the phosphorylation reaction of ADP that is linear for at least 60 sec and it can be calculated from the slope of the line. B. Linear regression analysis for the correlation between ATP amount and luciferin/luciferase-dependent luminescence. C. Example of the ATP synthetic activity (mean ± S.E.M.) using three preparations of digitonin-permeabilized HCT116 cells.
- Subtract the RLU at T0 from all other time points (Figure 2A).
Notes
- The concentration of digitonin and time of permeabilization are critical and might change depending on the cell line. Therefore, we recommend the previous titration of the digitonin concentration to be used in each cell line that better fits the determination of the ATP synthetic activity.
- It is recommended to prepare ADP freshly before use.
Recipes
- Respiration Medium (RM) (100 ml)
225 mM sucrose (MW = 342.2965 g/mol): 7.7 g
10 mM KCl (MW = 74.5 g/mol): 74 mg
5 mM MgCl2 (MW = 95.2 g/mol): 47.6 mg
0.05 % w/v bovine serum albumin: 50 mg
10 mM potassium-phosphate buffer [HK2PO4 (MW = 174.17 g/mol): 0.17 g; H2KPO4 (MW = 136.1 g/mol): 0.16 g]
1 mM EGTA (MW = 380.35 g/mol): 38 mg
10 mM Tris-HCl (pH 7.4) (MW = 121.14 g/mol): 38 mg - Mix A
Respiration Medium (RM)
0.1 mM ADP
5 mM succinate
0.15 µM P1, P5-di(adenosine-5′) pentaphosphate
2 µg/ml rotenone with or without 30 µM oligomycin - Mix B
RM
0.25 mg/ml of luciferin
0.02 mg/ml luciferase.
Materials and Reagents
- Microplate, 96 well, Polystyrene, black flat bottom, TC-treated (Cultek, catalog number: 153603 )
- Protein assay dye reagent concentrate (Bio-Rad Laboratories, catalog number: 5000006 )
- Liquid nitrogen
- Phosphatase inhibitor cocktail 2 (Sigma-Aldrich, catalog number: P5726 )
- cOmpleteTM, Mini, EDTA-free (Sigma-Aldrich, Roche Diagnostics®, catalog number: 11836170001 )
- Oligomycin (VWR International, catalog number: 80058-538 )
- Trizma® base (Sigma-Aldrich, catalog number: T1503 )
- Bovine serum albumin (Sigma-Aldrich, catalog number: A7906 )
- Adenosine 5′-triphosphate (ATP) disodium salt hydrate (Sigma-Aldrich, catalog number: FLAAS )
- KCl (Merck Millipore Corporation, catalog number: 104936 )
- MgCl2 (Merck Millipore Corporation, catalog number: 172571 )
- Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) (Sigma-Aldrich, catalog number: C2920 )
- Antimycin A (Sigma-Aldrich, catalog number: A8674 )
- Phospho(enol)pyruvic acid cyclohexylammonium salt (Sigma-Aldrich, catalog number: P3637 )
- L-Lactate dehydrogenase (L-LDH) from rabbit muscle (Sigma-Aldrich, Roche Diagnostics®, catalog number: 10127876001 )
- Pyruvate kinase from rabbit muscle (Sigma-Aldrich, catalog number: P9136 )
- β-Nicotinamide adenine dinucleotide, reduced dipotassium salt (NADH) (Sigma-Aldrich, catalog number: N4505 )
- Reaction Buffer (RB) stock solution (see Recipes)
- FCCP stock solution (see Recipes)
- Antimycin A stock solution (see Recipes)
- Complete reaction buffer (see Recipes)
- 10 mM PEP stock solution
- 2.5 mM ATP stock solution
- 1 mM NADH stock solution
- Oligomycin A stock solution
Equipment
- FLUOstar Omega Microplate Reader (BMG Labtech, model: FLUOstar Omega )
- Heraeus Megafuge 11R (Thermo Fisher Scientific, model: Heraeus Megafuge 11R )
- Centrifuge 5415R (Sigma-Aldrich, Eppendorf®, model: 5415R )
Procedure
- ATP hydrolysis by the enzyme can be determined spectrophotometrically following the changes in Absorbance at 340 nm (A340). The basis of this method is the coupling of the enzymatic reactions catalyzed by lactate dehydrogenase (LDH) and pyruvate kinase (PK) to the hydrolytic activity of the ATP synthase (Barrientos et al., 2009). The ADP generated by the hydrolysis of ATP allows PK to transform 1 mole of phosphoenolpyruvate (PEP) into 1 mole of pyruvate. Pyruvate is then used by LDH to generate lactate oxidizing a molecule of NADH to NAD+ in the process. By each ATP molecule hydrolyzed one molecule of NADH is oxidized, allowing the detection of the reaction by a diminution in A340.
- Determination of the hydrolytic activity of the H+-ATP synthase is assayed on isolated broken mitochondria by three cycles of freezing and thawing the mitochondrial preparation in liquid nitrogen and at 37 °C, respectively.
- 30-50 µg of isolated mitochondria are resuspended in 20 µl of Reaction Buffer (RB) with 1x phosphatase and protease inhibitors.
- The luminometer plate-reader is set up to measure absorbance at 340 nm in kinetic mode (10 flashes per well) and the change in the absorbance is measured through constant intervals of time (15 sec). With these conditions it is possible to measure 24 samples simultaneously.
- 80 µl of complete RB (containing PK and LDH) and 20 µl of the mitochondria prepared as on step 3 are added in each 96 well black clear bottom plate and the reaction is recorded.
- When enough ATP hydrolysis has been produced (2-5 min), 30 µM oligomycin is added to each well. The addition of oligomycin inhibits the activity of the mitochondrial ATP synthase/hydrolase providing confirmation of the specificity of the assay.
- The ATPase activity of isolated mitochondria is expressed in nanomoles of NADH oxidized/min/mg of mitochondrial protein which is calculated using the Lambert-Beer equation and a molar extinction coefficient of NADH of 6.22 x 103 M-1 cm-1.
- N = ΔA340/(ε340 x l) where N is the concentration of NADH oxidized, ΔA340 is the decrement in absorbance between two given time-points, ε340 is the Molar extinction coefficient of NADH at 340 nm and l is the light path length of 1 cm.
Representative data
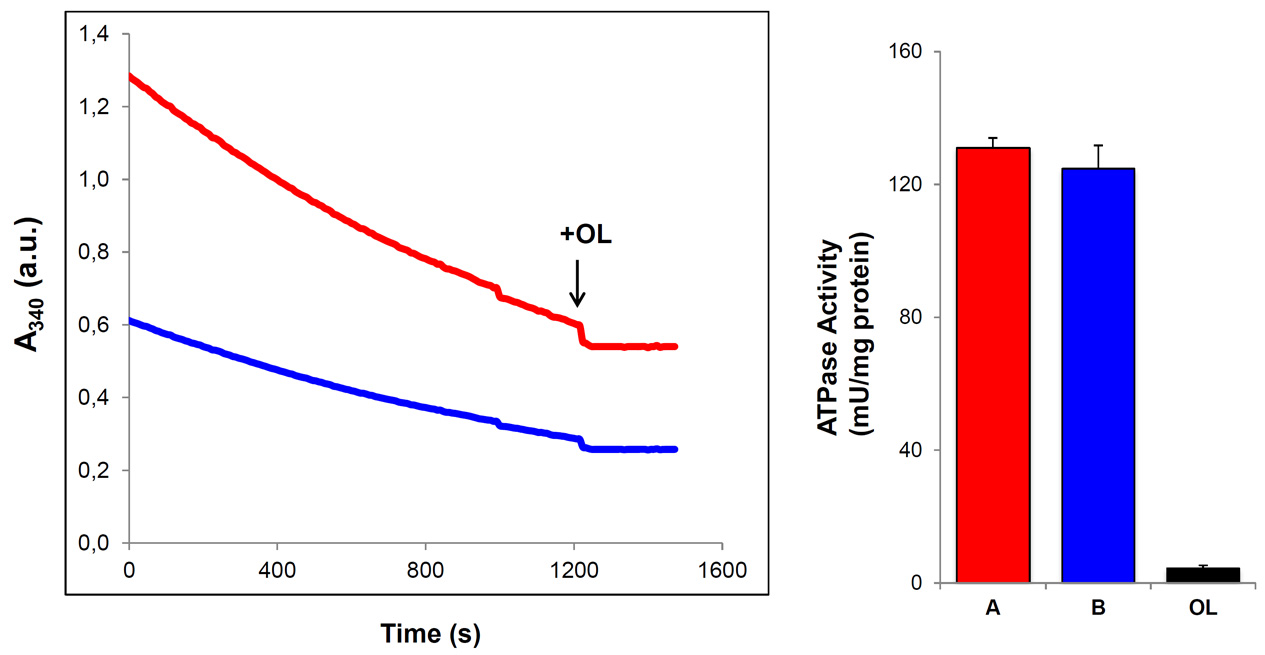
Figure 3. ATP hydrolase activity in isolated mitochondria from mouse heart. The slope of the graphs indicates the decrement of A340 as a function of reaction time. Where indicated, 30 μM of oligomycin (+OL, arrowhead) was added. Two different preparations containing 60 μg (A, red) and 30 μg (B, blue) of isolated mitochondria were assayed. Addition of OL (closed bar) blocks the hydrolase activity as evidence by the suppression of slope of the reaction. The ATP hydrolase activity is expressed as mU/mg protein. Bars represent the mean ± S.E.M. of the ATPase activity determined in three preparations of isolated mitochondria from mouse hearts.
Notes
- Note that the RB added at step 3 is not supplemented with PK and LDH.
- ATP and NADH must be prepared freshly.
- The FCCP, antimycin A, PEP and oligomycin stock solutions can be store at -20 °C.
Recipes
- Reaction Buffer (RB) stock solution (10 ml)
50 mM Tris-HCl (pH 8.0) (MW = 121.14 g/mol): 61 mg
5 mg/ml bovine serum albumin: 50 mg
20 mM MgCl2 (MW = 95.2 g/mol): 19 mg
50 mM KCl (MW = 74.55 g/mol): 37.2 mg - 2.5 mM FCCP stock solution
FCCP (MW = 254.17 g/mol)
Dissolve 0.6 mg FCCP in 1 ml of DMSO - 500 µM antimycin A stock solution
Antimycin A (MW = 548.63 g/mol)
Dissolve 0.3 mg antimycin A in 1 ml of ethanol - Complete Reaction Buffer, 1 ml (enough volume of RB to run ten assays)
- 0.92 ml RB stock solution
- 2 µl 2.5 mM FCCP stock solution (Final concentration: 5 μM)
- 2 µl 500 µM antimycin A stock solution (Final concentration: 1 μM)
- 10 µl 1 M PEP (Final concentration: 10 μM)
Dissolve 27 mg PEP in 100 μl of water - 25 µl 100 mM ATP (Final concentration: 2.5 mM)
Dissolve 5.5 mg in 100 µl of water - 10 µl 100 mM NADH (Final concentration: 1 mM)
Dissolve 7.4 mg in 100 µl of water
13 µl 4 units of LDH (2,750 U/ml)
20 µl 4 units of PK (1,920 U/ml)
Note: The addition of 4 units of LDH and of 4 units of PK is done in the last step and just before triggering the reaction by the addition of mitochondria. - 2 µl 1.5 mM Oligomycin A stock solution
Dissolve 1.2 mg oligomycin stock solution in 1 ml of ethanol
Acknowledgments
We thank Drs. María Sánchez-Aragó and Laura Formentini for expert guidance in setting the assays of the ATP synthase activities. The technical assistance of M. Chamorro and C. Nuñez de Arenas is acknowledged. The authors are grateful to the previous work developed in the labs of Drs. Enríquez, Barrientos and Manfredi for developing the methods adapted in these protocols. JGB and CNT were supported by pre-doctoral fellowships from FPI-MICINN/MINECO and Fondo Social Europeo, Spain. This work was supported by grants from Ministerio de Economía y Competitividad (SAF2013-41945-R), Comunidad de Madrid (S2011/BMD-2402), and Fundación Ramón Areces (FRA), Spain. The CBMSO receives an institutional grant from the FRA.
References
- Barrientos, A., Fontanesi, F. and Diaz, F. (2009). Evaluation of the mitochondrial respiratory chain and oxidative phosphorylation system using polarography and spectrophotometric enzyme assays. Curr Protoc Hum Genet Chapter 19: Unit19 13.
- Fernandez-Vizarra, E., Ferrin, G., Perez-Martos, A., Fernandez-Silva, P., Zeviani, M. and Enriquez, J. A. (2010). Isolation of mitochondria for biogenetical studies: An update. Mitochondrion 10(3): 253-262.
- Vives-Bauza, C., Yang, L. and Manfredi, G. (2007). Assay of mitochondrial ATP synthesis in animal cells and tissues. Methods Cell Biol 80: 155-171.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
García-Bermúdez, J., Nuevo-Tapioles, C. and Cuezva, J. M. (2016). Determination of the H+-ATP Synthase and Hydrolytic Activities. Bio-protocol 6(16): e1905. DOI: 10.21769/BioProtoc.1905.
Category
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











