- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Modified Chromogenic Assay for Determination of the Ratio of Free Intracellular NAD+/NADH in Streptococcus mutans
Published: Vol 6, Iss 16, Aug 20, 2016 DOI: 10.21769/BioProtoc.1902 Views: 13869
Reviewed by: Valentine V TrotterSwetha ReddyRolf Lood

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

β-lactamase (Bla) Reporter-based System to Study Flagellar Type 3 Secretion in Salmonella
Fabienne F. V. Chevance and Kelly T. Hughes
Jun 20, 2023 1774 Views

Determination of Poly(3-hydroxybutyrate) Content in Cyanobacterium Synechocystis sp. PCC 6803 Using Acid Hydrolysis Followed by High-performance Liquid Chromatography
Janine Kaewbai-ngam [...] Tanakarn Monshupanee
Aug 20, 2023 1824 Views

An HPLC-based Assay to Study the Activity of Cyclic Diadenosine Monophosphate (C-di-AMP) Synthase DisA from Mycobacterium smegmatis
Avisek Mahapa [...] Dipankar Chatterji
Dec 20, 2024 1789 Views
Abstract
Nicotinamide adenine dinucleotide is a coenzyme present in all kingdoms of life and exists in two forms: oxidized (NAD+) and reduced (NADH). NAD(H) is involved in a multitude of essential metabolic redox reactions, providing oxidizing or reducing equivalents. The ratio of free intracellular NAD+/NADH is fundamentally important in the maintenance of cellular redox homeostasis (Ying, 2008). Various chromogenic cycling assays have been used to determine the ratio of NAD+/NADH in both bacterial and mammalian cells for more than forty years (Bernofsky and Swan, 1973; Nisselbaum and Green, 1969).
Here, we describe in detail an assay to determine the ratio of free intracellular NAD+ to NADH in Streptococcus mutans. This cycling assay is a modified version of the protocol first described by Bernofsky and Swan (Bernofsky and Swan, 1973), using the extraction buffer described by Frezza et al. (2011), followed by the reduced MTT precipitation described by Gibbon and Larher (Gibon and Larher, 1997). As depicted in Figure 1, alcohol dehydrogenase is used to drive a series of redox reactions utilizing exogenously added ethanol and NAD+ from sample extracts as initial substrates, phenazine ethosulfate (PES) as an electron carrier, and thiazolyl blue tetrazolium bromide (MTT) as a terminal electron acceptor. 6 M NaCl is used to stop the reaction. The reduced MTT (formazan dye) is purple in color and can be quantified by measuring absorbance at 570 nm. This protocol is divided into three steps: A. Preparation of cell pellets of S. mutans; B. Preparation of deproteinated cell extracts containing NADtotal or NADH; C. NAD+/NADH cycling assay. This method has proven robust in measuring the NAD+/NADH ratio in S. mutans under a variety of conditions, and should be applicable to other Gram-positive bacteria.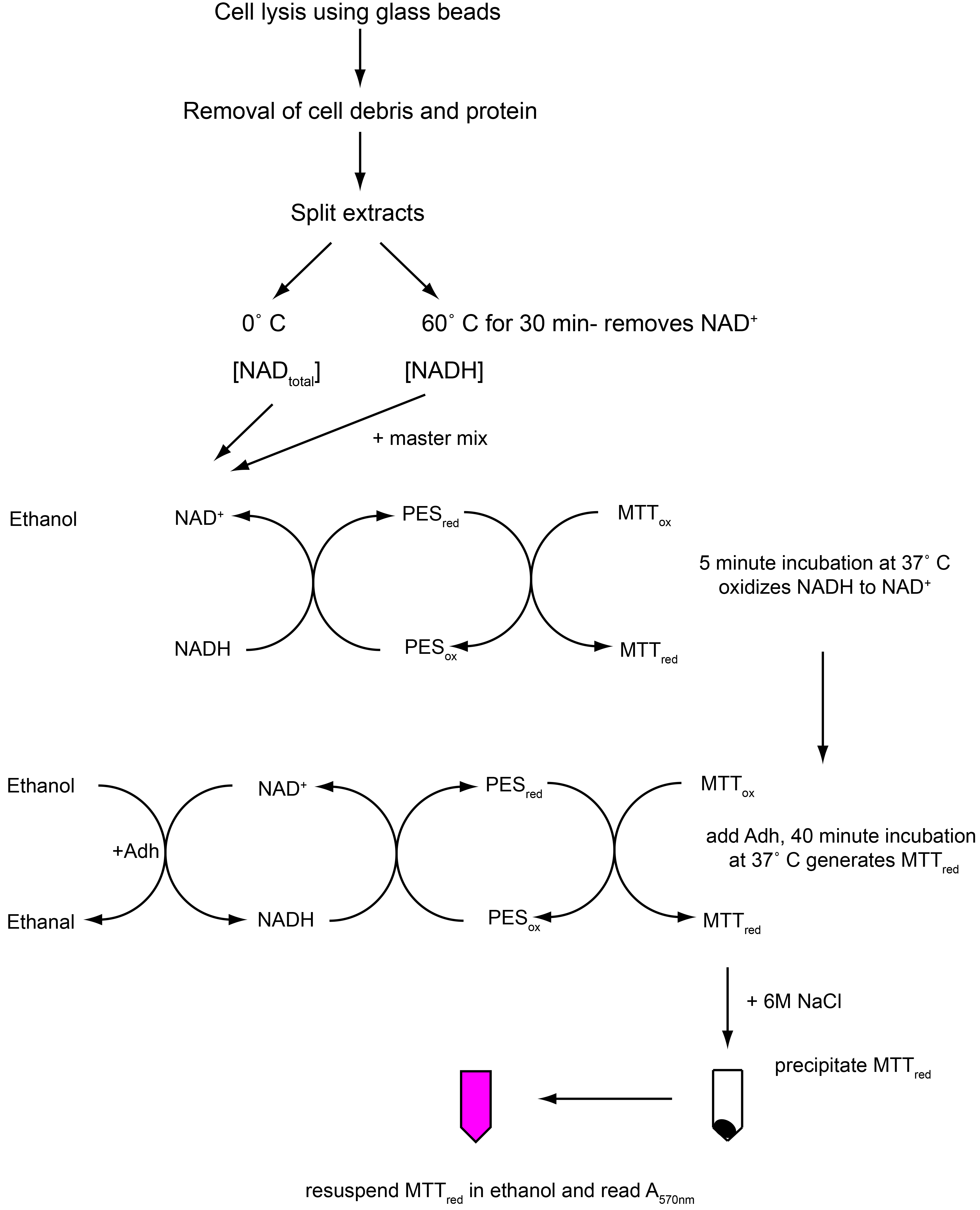
Figure 1. Flowchart illustrating protocol Procedure parts B-C
Materials and Reagents
- Glass test tubes, 18 x 150 mm (VWR International, catalog number: 60825-443 )
- 50 ml centrifuge tubes (Corning, catalog number: 430829 )
- Screw-top microcentrifuge tubes with O-ring, 2.0 ml (Laboratory Product Sales, catalog number: L233072 )
- Glass beads, 0.1 mm (BioSpec Products, catalog number: 11079101 )
- Centrifugal Filters, 0.5 ml, 10,000 MWCO (Merck Millipore Corporation, Amicon, catalog number: UFC501024 )
- Microcentrifuge tubes, 1.7 ml (Laboratory Product Sales, catalog number: L211511 )
- Streptococcus mutans bacterial strains of interest
- Brain Heart Infusion medium (BHI) (BD, DifcoTM, catalog number: 299070 )
- Bacto-agar (BD, DifcoTM, catalog number: 214010 )
- Tricine (Sigma-Aldrich, catalog number: T5816 )
- Sodium hydroxide (Sigma-Aldrich, catalog number: S5881 )
- Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: E5134 )
- Sodium chloride (J.T. Baker, catalog number: 3624-01 )
- Thiazolyl blue tetrazolium bromide (MTT) (Sigma-Aldrich, catalog number: M2128 )
- Phenazine ethosulfate (PES) (Sigma-Aldrich, catalog number: P4544 )
- Alcohol dehydrogenase from Saccharomyces cerevisiae (Adh) (Sigma-Aldrich, catalog number: A3263 )
- Ethanol, 200 proof (VWR International, Koptec, catalog number: 64175 )
- Sodium bicarbonate (Sigma-Aldrich, catalog number: S8875 )
- Sodium carbonate (J.T. Baker, catalog number: 3602-01 )
- Triton X-100 (Sigma-Aldrich, catalog number: T8787 )
- Nicotinamide (Sigma-Aldrich, catalog number: N3376 )
- Hydrochloric acid (J.T. Baker, catalog number: 9535 )
- β-Nicotinamide adenine dinucleotide, reduced disodium salt hydrate (NADH) (Sigma-Aldrich, catalog number: N8129 )
- 1 M tricine-NaOH (see Recipes)
- 40 mM EDTA solution (see Recipes)
- 6 M NaCl (see Recipes)
- 0.1 M NaCl (see Recipes)
- 4.2 mM MTT (see Recipes)
- 16.6 mM PES (see Recipes)
- 100 U/ml Adh (see Recipes)
- NAD+/NADH master mix (see Recipes)
- NAD+/NADH extraction buffer (see Recipes)
- 2 µM NADH (see Recipes)
Equipment
- 3 mm (1 µl) inoculation loops (VWR, catalog number: 50807-020 )
- CO2 incubator (LABEQUIP) (VWR Scientific, catalog number: 10810-744 )
- Erlenmeyer flasks, 250 ml (Pyrex)
- Mini Beadbeater 8 (BioSpec Products, model: Mini Beadbeater 8 )
- Microcentrifuge (Eppendorf, model: 5424 )
- Centrifuge (Fisher Scientific, model: Legend RT) with Thermo swinging bucket rotor (Thermo Fisher Scientific, model: TX-750 )
- 60 °C water bath
- 37 °C water bath
- Vortexer (Vortex Genie 2) (Scientific Industries, model: G560 )
- Dark room
Procedure
- Preparation of S. mutans cell pellets (Note 1)
- The S. mutans bacterial strains of interest were streaked for isolation from frozen culture stocks, on BHI plates (37 g/L BHI; 15 g/L Bacto agar) using 1 µl loops and incubated overnight at 37 °C in a 5% (v/v) CO2/95% air incubator.
- Using sterile technique, an isolated colony was picked with a 1 µl loop and used to inoculate 5 ml BHI in a sterile glass test tube. Cultures were grown overnight 37 °C in a 5% (v/v) CO2/95% air incubator (Note 2).
- Using sterile technique, the 5 ml overnight culture was used to inoculate 45 ml BHI in a sterile 125 ml culture flask. Cultures were grown at 37 °C in a 5% (v/v) CO2/95% air incubator until the absorbance of the culture at 600 nm (OD600) reached ~0.6 (mid-log phase growth, roughly 4 h for type strain UA159).
- Cultures were transferred to 50 ml conical centrifuge tubes and cells were harvested by centrifugation at 4,000 rpm (2,272 x g) for 15 min at 4 °C. Cell pellets were placed at -80 °C until the assays were performed.
- The S. mutans bacterial strains of interest were streaked for isolation from frozen culture stocks, on BHI plates (37 g/L BHI; 15 g/L Bacto agar) using 1 µl loops and incubated overnight at 37 °C in a 5% (v/v) CO2/95% air incubator.
- Preparation of cell extracts from S. mutans
- Frozen cell pellets were thawed on ice and resuspended in 1 ml ice-cold NAD+/NADH extraction buffer (Frezza et al., 2011).
- Samples were transferred into 2.0 ml screw-capped microcentrifuge tubes containing 0.5 g glass beads.
- Samples were homogenized twice in a Mini Beadbeater on the “homogenize” speed setting at 4 °C for a 30 sec cycle, followed by a 2 min incubation on ice, then another 30 sec cycle.
- Homogenized samples were centrifuged at 4 °C in a microcentrifuge at 14,000 x g for 5 min.
- Supernatants were removed and transferred to a 10,000 MWCO centrifugal filter (Note 3).
- Supernatant was passed through the filter by centrifugation at 14,000 x g at 4 °C. With the sample size used, the filtration typically took about 30 min.
- The filtrate was split into two equal volumes in fresh microcentrifuge tubes (~400 µl each), one for determination of total NAD/NADH (NADtotal), and one for determination of NADH. From this point until step C7, the protocol must be carried out in a dark room (Note 4).
- The sample aliquot for NADH only was incubated in a 60 °C water bath for 30 min to remove NAD+ then transferred to ice (NAD+ is heat labile, NADH is not). The aliquot for determination of NADtotal was maintained on ice throughout this step (Note 5).
- Frozen cell pellets were thawed on ice and resuspended in 1 ml ice-cold NAD+/NADH extraction buffer (Frezza et al., 2011).
- NAD+/NADH cycling assay
- Aliquot 500 µl of master mix (Gibon and Larher, 1997) into an appropriate number of microcentrifuge tubes (Note 5).
- Add 100 µl of sample, NADH standard, or NAD+/NADH extraction buffer (for blank) to each microcentrifuge tube containing master mix.
- Bring each reaction volume to 900 µl with 0.1 M NaCl and vortex briefly.
- Incubate reactions in a 37 °C water bath for 5 min (Note 6).
- Add 100 µl of Adh (containing 10 U) to each reaction and vortex briefly.
- Incubate reactions in a 37 °C water bath for 40 min.
- Add 500 µl of 6 M NaCl to stop each reaction and precipitate the reduced MTT. Vortex briefly. The reactions are no longer light sensitive and the remainder of the protocol may be carried out under standard lighting conditions.
- Centrifuge stopped reactions at 4 °C and 10,000 x g for 5 min.
- Decant tubes (Note 7) and resuspend pellets in 1 ml 200 proof ethanol.
- Analyze samples and standards for absorbance at 570 nm, using the sample containing 100 µl NAD+/NADH extraction buffer as a blank. Generate a standard curve using known quantities of NADH (added in step C2 above) and use to calculate the concentration of unknown samples. Calculate NAD+ via NADtotal-NADH.
- Aliquot 500 µl of master mix (Gibon and Larher, 1997) into an appropriate number of microcentrifuge tubes (Note 5).
Notes
- Part A of this protocol describes preparing S. mutans cells grown in batch culture, however the authors expect the downstream portion of the protocol (Part B and further) to function for any Gram-positive bacterial species and growth condition. In addition to S. mutans batch cultures, the authors have also successfully performed the assay on samples of S. mutans grown in continuous cultures.
- As a facultative anaerobe, Streptococcus mutans is typically grown without agitation.
- This step removes cell debris and protein from the cell extracts, preventing sequestration, degradation, oxidation, or reduction of the NAD(H).
- From this point forward until step C7, the protocol must be performed in a dark room, with only an indirect light source to prevent oxidation of NAD(H), MTT, and PES.
- The NAD+/NADH master mix and NADH standards were prepared fresh for every assay as described in Recipes during the 30 min incubation described in step B8. An assay should be performed on several dilutions of each sample to ensure that the resulting absorbance values fall in within the linear range of the generated standard curve. A representative standard curve is displayed in Figure 2. The authors used solutions of 0 pmol, 10 pmol, 25 pmol, 50 pmol, 100 pmol, and 200 pmol to generate the standard curve. As there is generally a greater proportion of free intracellular NAD+ than NADH, it is likely the NADtotal samples will need to be diluted to a greater extent than the NADH samples. In addition to 100 µl of non-diluted sample, the authors recommend performing assays on 100 µl of 1:5, 1:10, and 1:50 dilutions of NADtotal samples and 100 µl of 1:2, 1:5 and 1:10 dilutions for NADH samples. For each sample and standard, three replicate assays are performed. A negative control reaction must also be performed (to use as a blank in the spectrophotometer) using 100 µl NAD+/NADH extraction buffer.
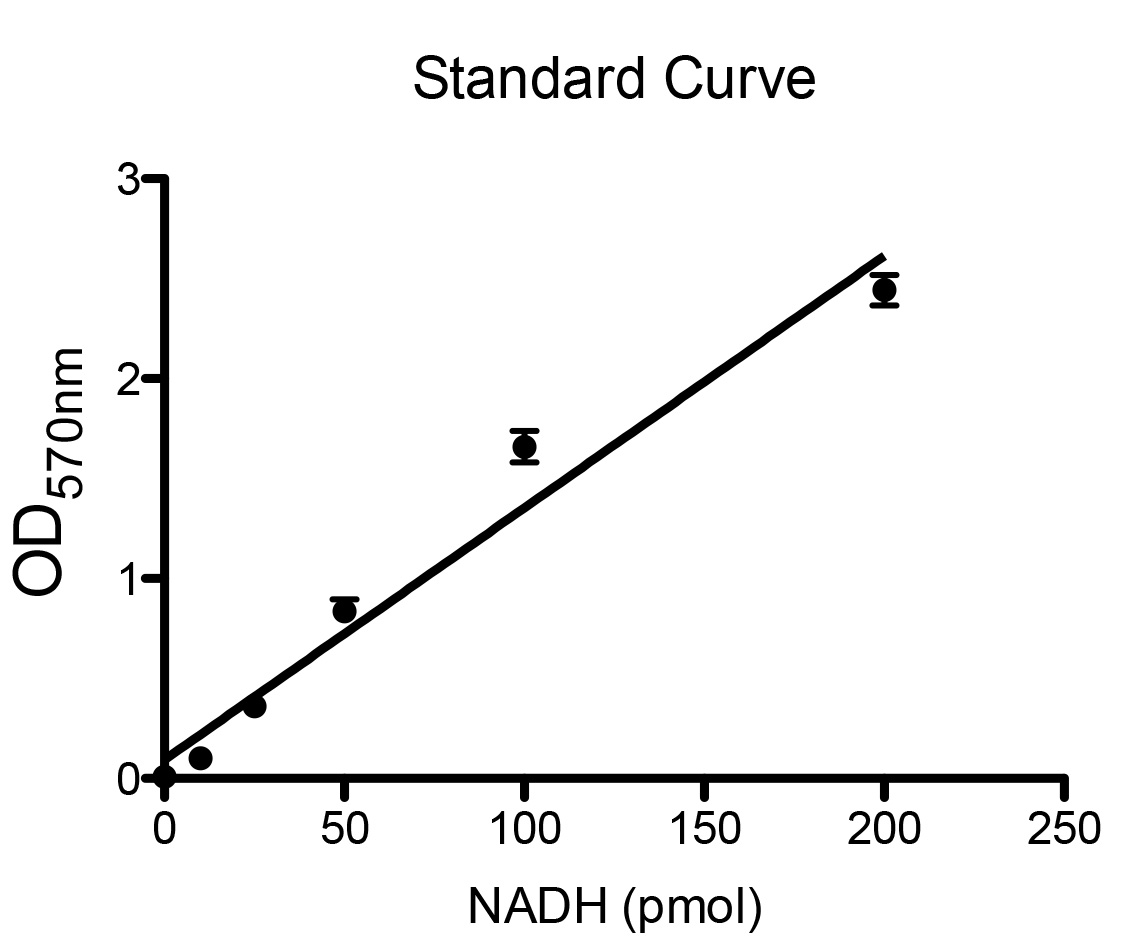
Figure 2. Example of standard curve generated by several known concentrations of NADH - Incubation at 37 °C results in oxidization of all the NADH into NAD+.
- Samples are no longer highly light-sensitive following this step and the remainder of the protocol can be performed in standard lighting conditions.
Recipes
- 1 M tricine-NaOH (pH 8)
Add 17.92 g tricine to 80 ml H2O
Adjust pH to 8.0 with NaOH
Bring volume of solution to 100 ml with H2O. Store at RT. - 40 mM EDTA
Add 1.49 g EDTA to 80 ml H2O
Bring volume of solution to 100 ml with H2O. Store at RT. - 6 M NaCl
Add 35.04 g NaCl to 80 ml H2O
Bring volume of solution to 100 ml with H2O. Store at RT. - 0.1 M NaCl
Add 0.584 g NaCl to 80 ml H2O
Bring volume of solution to 100 ml with H2O. Store at RT. - 4.2 mM MTT
Add 0.0087 g MTT to 5 ml H2O
Prepare fresh each time the protocol is performed and keep in the dark. - 16.6 mM PES
Add 0.028 g PES to 5 ml H2O
Prepare fresh each time the protocol is performed and keep in the dark. - 100 U/ml Adh
Add appropriate amount of Adh powder to 0.1 M tricine-NaOH (pH 8) to get 100 U/ml (units of activity per mg solid can be calculated from provided total units and mg solid provided on the label of each tube of enzyme).
Prepare fresh each time the protocol is performed. - NAD+/NADH master mix
Prepare fresh each time the protocol is performed.
Prepare a volume based upon the number of reactions to be performed (500 µl for each reaction).
Must be mixed and stored in the dark.
Per reaction add:
100 µl 1 M tricine-NaOH (pH 8)
100 µl 40 mM EDTA
100 µl 0.1 M NaCl
100 µl 4.2 mM MTT
100 µl 16.6 mM PES
100 µl 200 proof ethanol - NAD+/NADH extraction buffer pH 10.3 (100 ml)
To 80 ml H2O add:
0.168 g sodium bicarbonate (final concentration of 20 mM)
1.06 g sodium carbonate (final concentration of 100 mM)
50 µl Triton-X 100 (final concentration of 0.05%)
0.122 g nicotinamide (final concentration of 10 mM)
After reagents are dissolved, adjust pH to 10.3 with 1 M HCl.
Bring to 100 ml final volume.
Store at RT. - 2 µM NADH (for standard curve preparation)
Add 0.007 mg to 5 ml H2O
This quantity results in a 1,000x solution, so 3 serial dilutions (100 µl into 900 µl H2O) are necessary in order to get a 1x working stock.
Standard curve
0 pmol NADH = 1 ml H2O
10 pmol NADH = 5 µl of 2 µM NADH into 995 µl H2O
25 pmol NADH = 12.5 µl of 2 µM NADH into 987.5 µl H2O
50 pmol NADH = 25 µl of 2 µM NADH into 975 µl H2O
100 pmol NADH = 50 µl of 2 µM NADH into 950 µl H2O
200 pmol NADH = 100 µl of 2 µM NADH into 900 µl H2O
Acknowledgments
This work was supported by the Training Program in Oral Sciences, NIH/NIDCR T90-DE021985 (J. L. B.), and by NIH/NIDCR DE-13683 (R. G. Q), NIH/NIDCR DE-17425 (R. G. Q.).
This cycling assay is a modified version of the protocol first described by Bernofsky and Swan (1973), using the extraction buffer described by Frezza et al. (2011), followed by the reduced MTT precipitation described by Gibbon and Larher (1977).
References
- Bernofsky, C. and Swan, M. (1973). An improved cycling assay for nicotinamide adenine dinucleotide. Anal Biochem 53(2): 452-458.
- Frezza, C., Zheng, L., Tennant, D. A., Papkovsky, D. B., Hedley, B. A., Kalna, G., Watson, D. G. and Gottlieb, E. (2011). Metabolic profiling of hypoxic cells revealed a catabolic signature required for cell survival. PLoS One 6(9): e24411.
- Gibon, Y. and Larher, F. (1997). Cycling assay for nicotinamide adenine dinucleotides: NaCl precipitation and ethanol solubilization of the reduced tetrazolium. Anal Biochem 251(2): 153-157.
- Nisselbaum, J. S. and Green, S. (1969). A simple ultramicro method for determination of pyridine nucleotides in tissues. Anal Biochem 27(2): 212-217.
- Ying, W. (2008). NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxid Redox Signal 10(2): 179-206.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Baker, J. L., Faustoferri, R. C. and Quivey, Jr, R. G. (2016). A Modified Chromogenic Assay for Determination of the Ratio of Free Intracellular NAD+/NADH in Streptococcus mutans . Bio-protocol 6(16): e1902. DOI: 10.21769/BioProtoc.1902.
Category
Microbiology > Microbial metabolism > Other compound
Microbiology > Microbial biochemistry > Other compound
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










