- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Capillary Electrophoresis in Hydroxyethylcellulose Solutions for the Analysis of dsDNA, dsRNA, and siRNA
Published: Vol 6, Iss 15, Aug 5, 2016 DOI: 10.21769/BioProtoc.1894 Views: 9917
Reviewed by: Samik BhattacharyaAmit DeyAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Methylation-sensitive Amplified Polymorphism as a Tool to Analyze Wild Potato Hybrids
Nicolás Cara [...] Ricardo W. Masuelli
Jul 5, 2020 5231 Views

Electrophoretic Mobility Assay to Separate Supercoiled, Catenated, and Knotted DNA Molecules
Jorge Cebrián [...] María José Fernández-Nestosa
May 5, 2024 2152 Views

Simple Analysis of Gel Images With IOCBIO Gel Software
Lucia Jaska [...] Marko Vendelin
Aug 20, 2024 1629 Views
Abstract
Capillary electrophoresis (CE) is identified as a promising technology for the study of nucleic acid molecules because of its high efficiency, high throughput with automation and integration. Compared to the traditional method of slab gel electrophoresis (SGE), the advantages of CE cannot be emphasized more. Most of CE process, including sample injection, detection and data analysis, is able to be automated which will save great labor for industrial and research labs. CE used the separation channel with micrometer-scale diameter, so the joule heat is easy to be dissipated during electrophoresis. Thus high separation voltage (> 100 V/cm) is allowed in CE while in SGE (usually ~10 V/cm) it usually causes severe band broadening. Because the band broadening is restrained efficiently in CE, it is capable of detecting minute samples and becoming more sensitive than SGE. The advantage of allowing high voltage consequently speeds up the CE separation and yields a better throughput compared to SGE. CE costs less reagents, for example buffer solutions, sieving matrix, dye reagents etc. In addition, the micrometer-scale channel is easy to be integrated with upstream and downstream sample treatment units, forming a lab on a chip. This merit of CE already attracted considerable interests among researchers from various areas.
The difficulties of CE involve filling the gels (agarose or cross-linked polyacrylamide) into the capillary tube. Also, the reproducibility and the life-time of the gel-capillary are limited. But the small-diameter capillary allows to use replaceable polymer solutions, which can efficiently prevent the convection of the separation buffer. Polymer solutions are easier to be filled into the capillary and yield more stable separations. Thus, those difficulties are resolved by doing capillary polymer electrophoresis (CPE), which is going to be described in this protocol.
Several separation modes, for example, capillary gel electrophoresis (CGE), CPE, capillary zone electrophoresis (CZE), capillary isotachophoresis (CITP) and so on, have been developed for analysis of different kinds of molecules. Here, we introduce the protocol for CPE in detail, which is for the separation of dsDNA, dsRNA (including siRNA) molecules. Polymer solutions are filled into the capillary as a sieving matrix for double strand nucleic acids separation. Hydroxyethylcellulose (HEC) polymer is employed as the sieving polymer in this case. A home-built CE system is described in detail.
Materials and Reagents
- Micro-centrifuge or PCR tube (200 μl)
- Pipettes (0.1-10 μl and 10-200 μl) and the corresponding tips
- 0.22 μm filter membrane
- dsDNA analytes (Takara, catalog number: 3420A )
- dsRNA (including siRNA) analytes (Takara, catalog number: 3430 )
- Ultra-pure water, with resistance of 18.25 Ω
- 2-hydroxyethyl cellulose (MW: 1,300k) (Sigma-Aldrich, catalog number: 434981 )
- TBE (Tris-borate-EDTA) powder (Takara, catalog number: T905 )
- SYBR Green II RNA Gel Stain, 10,000x concentrate in DMSO (Thermo Fisher Scientific, catalog number: S-7564 )
- Storage 1% HEC polymer solution (see Recipes)
- 0.5x TBE solution (see Recipes)
- 100x SYBR Green II (see Recipes)
Equipment
- Home-built CE system (Figure 1) which includes:
- A coated fused silica capillary with a detection window (Polymicro, catalog number: TSP075375 )
- A high voltage power supplier (see Note 8)
- A fluorescence microscope assembled with a fluorescent excitation equipment (Equipment 1d) and a fluorescent emission collection equipment (Equipment 1e) (Olympus, model: IX73 )
- The fluorescent excitation equipment constituted by a mercury lamp and an optical cube, which was used to transmit light from the mercury lamp into 460-495 nm fluorescence excitation spectrum (Olympus, model: U-MWIB3 )
- The fluorescent emission collection equipment constituted by an optical filter and a photomultiplier tube (PMT) (Hamamatsu Photonics, catalog number: H8249-101 , for alternate H7827-001)
Note: The optical filter transmits the emission spectrum from dye-nucleonic acid conjugate to the PMT and filters background light from the detection window. - The home-built CE system including a Labview system (National Instrument, model: NI USB 6212 ) and a computer
Note: Users are able to control the high voltage supplier, monitor fluorescent signal from the detection window and collect data from PMT by the LabVIEW software. - Vacuum pump (see Note 9)
- Centrifuge (> 3,200 x g)
- Ultra-pure water system (see Note 10)

Figure 1. Schematic diagram of the assembly of the CE system
Software
- LabVIEW software
Procedure
- Prepare CE sample. Collect the dsDNA/dsRNA analytes into a micro-centrifuge tube, for example, a 200 μl volume micro-centrifuge tube. Dissolve or dilute the dsDNA/dsRNA analytes with 0.5x TBE (see Note 3).
- Prepare 0.5% HEC polymer solution (see Note 4). Take one micro-centrifuge tube, pipet into 100 μl storage 1% HEC polymer solution, then add into 96 μl 0.5x TBE solution and 4 μl 100x SYBR Green II. Mix the buffer solution by pipetting for several times. Split the 0.5% HEC polymer solution in another micro-centrifuge tube. Centrifuge the micro-centrifuge tubes for 5-20 sec so that the bubbles are removed from the buffer solution. In this case, we take the 0.5% HCE for an example. The concentration of polymer solution affects the separation performance of CPE greatly (Liu et al., 2015). Users are suggested to adjust the concentration of polymer solution according to the sample to be analyzed.
- Clean the inner wall of the capillary (Figure 2). Flush the capillary by ultra-pure water for ~30 sec using the vacuum pump. The sample will be injected at the cathode end of the capillary. In order to avoid inappropriate sample injections, we suggest placing the vacuum pump at the anode end of the capillary.
- Fill the 0.5% HEC polymer solution into capillary (Figure 2) as the same way of step 3.
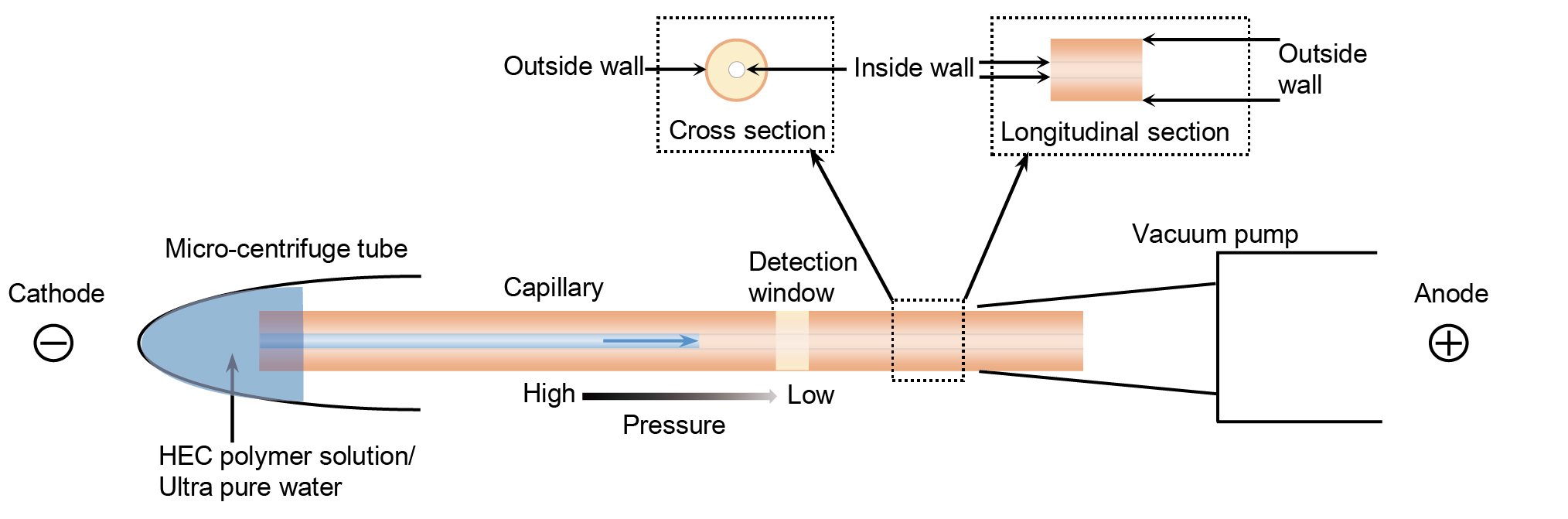
Figure 2. Schematic plot of filling the 0.5% HEC polymer solution into capillary and cleaning the inner wall of the capillary
- Set up CE parameters on the Labview interface: sample loading voltage, sample loading time, electric field intensity. When different samples are to be analyzed, those parameters are supposed to be optimized to achieve preferable separation.
- Inject sample. As shown in Figure 3, immerse the cathode of power supplier and the corresponding end of the capillary into the sample solution in a micro-centrifuge tube. Immerse the anode of power supplier and the corresponding end of the capillary into a micro-centrifuge tube which contained 0.5% HEC polymer solution. Make sure the capillary is well connected with the power supplier. Through the LabVIEW control software installed on the computer, set on the high voltage power supplier. So that the high voltage is applied on the capillary and the sample is injected electrokinetically.

Figure 3. Schematic plot of injecting sample - Start CE separation. Remove the sample micro-centrifuge tube and place a micro-centrifuge tube which contained 0.5% HEC polymer solution in the same way. Make sure the capillary is well connected with the power supplier.
- The fluorescent signal can be monitored by the LabVIEW software during the CE separation process. In the first CE run, when the last signal is observed which is supposed from the last analyte, keep the CE process on for another same duration time in order to ensure no other signals appear. Save CE data. Proceed data analysis.
- Clean the inner wall of capillary following step 3. The clean-up prevents the next CPE run from contamination. If no more CPE run needed, it helps to maintain the capillary for long-term storage.
Representative data
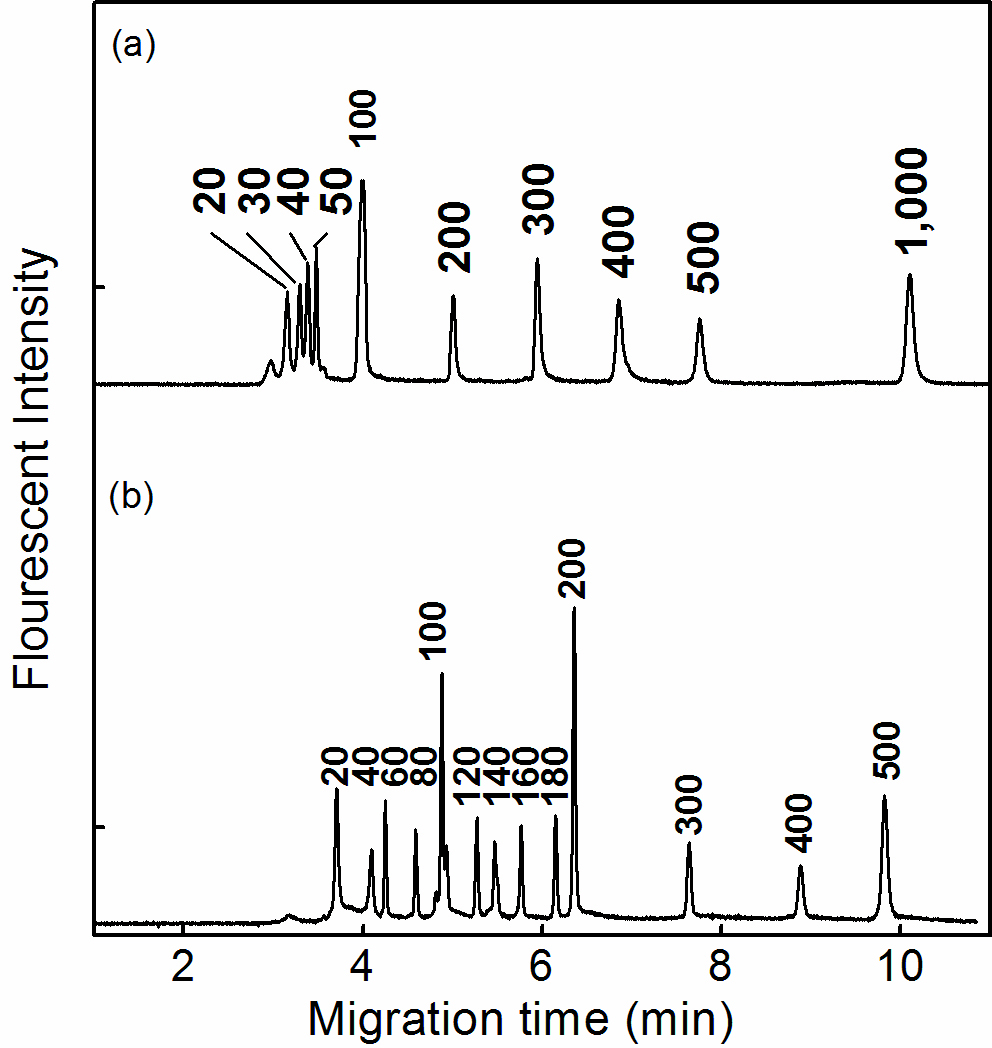
Figure 4. Electropherogram of (a) dsRNA fragments and (b) dsDNA fragments in 1.2% HEC (MW: 1,300k) solution. Capillary electrophoresis was performed at 100V/cm. The total length and the effective length of the capillary were 9.0 cm and 6.0 cm, respectively. The sample was loaded at 100 V/cm for 2.0 sec. The dsDNA sample and dsRNA sample was diluted into 13 ng/μl and 20 ng/μl, respectively, using 0.5x TBE.
Notes
- Denature is not obligatory for electrophoretic separation of dsDNA and dsRNA molecules.
- You can keep dsDNA and dsRNA analytes at 4 °C for short-term storage, and keep them at -20 °C for long term storage.
- Analytes are suggested to be resolved into 0.5x TBE in order to ensure the electrokinetic injection is the same for every time.
- Because the polymer solutions are viscous and take a long time to get resolved homogenous, it is recommended to prepare a concentrated polymer solution for storage. Before CPE experiment, the concentrated polymer solution can be diluted to a required concentration conveniently. In this case, we demonstrated the protocol to prepare the 0.5% HEC polymer solution with a final concentration of 0.5x TBE and 2x SYBR Green II.
- HEC polymers in various MWs, for example 90k-1,300k, can be used for dsDNA and dsRNA analysis. If the MW of HEC polymer is low, high concentration is recommended for dsDNA and dsRNA separation.
- SYBR Green I can also be used for dsDNA and dsRNA dying.
- It is suggested to keep capillaries in ultra-pure water for storage in order to maintain the performance of coated surface.
- Please make sure that the ripple of high voltage should be less than 0.1% of the applied voltage, and the slew rate for high voltage should be more than 20 V/μsec. These parameters are usually written in the product specification.
- If the capillary is long, a high pump pressure or a syringe is recommended in order to fill viscous polymer solution into the capillary. If the capillary is short, low pressure is preferred in case bubble formed inside the capillary. In our lab, the common capillary length is no more than 20 cm. The ultimate pressure of our vacuum pump is 26.6 x 103 Pa.
- The ultra-pure water system is supposed to produce water which meets ASTM (American Society of Testing Materials) Type-1.1 standard.
- We believe that the commercial available CE system is compatible with this protocol with an accommodation of the experimental process.
Recipes
- Storage 1% HEC polymer solution, 10 ml
1 g HEC powder
9 ml 0.5x TBE
Stir the solution at room temperature for at least 24 h in order to achieve a homogenous polymer solution. Keep storage polymer solutions at room temperature. - 0.5x TBE 1,000 ml
50 ml 10x TBE
950 ml ultra-pure water
Filter the 0.5x TBE solution twice using a 0.22 μm filter membrane.
Keep at room temperature. - 100x SYBR Green
1 μl 10,000x SYBR Green
99 μl ultra-pure water
Keep at 2-8 °C for at least one month.
Acknowledgments
This work was adapted and modified from Yamaguchi and Liu et al., 2015.
This project was partly supported by Grand-in-Aid for Scientific Research [No. 25600049 (Y. Y.), A15H038270 (Y. Y)], JSPS, Japan. We acknowledged on partly financial support by East China University of Science and Technology (No. YK0142119).
References
- Li, Z., Dou, X., Ni, Y., Chen, Q., Cheng, S. and Yamaguchi, Y. (2012). Is pulsed electric field still effective for RNA separation in capillary electrophoresis? J Chromatogr A 1229: 274-279.
- Liu, C., Yamaguchi, Y., Zhu, X., Li, Z., Ni, Y. and Dou, X. (2015). Analysis of small interfering RNA by capillary electrophoresis in hydroxyethylcellulose solutions. Electrophoresis 36(14): 1651-1657.
- Sumitomo, K., Sasaki, M. and Yamaguchi, Y. (2009). Acetic acid denaturing for RNA capillary polymer electrophoresis. Electrophoresis 30(9): 1538-1543.
- Todorov, T. I., Yamaguchi, Y. and Morris, M. D. (2003). Effect of urea on the polymer buffer solutions used for the electrophoretic separations of nucleic acids. Anal Chem 75(8): 1837-1843.
- Yamaguchi, Y., Todorov, T. I., Morris, M. D. and Larson, R. G. (2004). Distribution of single DNA molecule electrophoretic mobilities in semidilute and dilute hydroxyethylcellulose solutions. Electrophoresis 25(7-8): 999-1006.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Liu, C., Yamaguchi, Y. and Dou, X. (2016). Capillary Electrophoresis in Hydroxyethylcellulose Solutions for the Analysis of dsDNA, dsRNA, and siRNA. Bio-protocol 6(15): e1894. DOI: 10.21769/BioProtoc.1894.
Category
Molecular Biology > DNA > Electrophoresis
Molecular Biology > RNA > Electrophoresis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









