- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Generation of IgG-Fc Glycovariants Using Recombinant Glycosidases and Glycosyltransferases
Published: Vol 6, Iss 15, Aug 5, 2016 DOI: 10.21769/BioProtoc.1886 Views: 13281
Reviewed by: Jia LiOmar AkilElizabeth V. Clarke

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Proximity Labelling to Quantify Kv7.4 and Dynein Protein Interaction in Freshly Isolated Rat Vascular Smooth Muscle Cells
Jennifer van der Horst and Thomas A. Jepps
Mar 20, 2024 1569 Views

Determination of Ligand-Target Interaction in vitro by Cellular Thermal Shift Assay and Isothermal Dose-response Fingerprint Assay
Danyu Du [...] Jing Xiong
Aug 5, 2024 4963 Views

Ub-POD: A Ubiquitin-Specific Proximity-Dependent Labeling Technique to Identify E3 Ubiquitin Ligase Substrates in Human Cells
Urbi Mukhopadhyay [...] Sagar Bhogaraju
Jun 20, 2025 2458 Views
Abstract
The immunoglobulin G (IgG) fragment crystallizable (Fc) domain contains a single, highly conserved asparagine 297 (N297) glycosylation site in the CH2 domain, which is buried within the hydrophobic core of each of the two heavy chains. The biantennary core glycan structure, composed of 2 N-acetylglucosamine (GlcNAc) and 3 mannose residues, can be further decorated with fucose, bisecting GlcNAc and terminal GlcNAc, galactose, and sialic acid. Presence or absence of distinct residues can alter IgG effector functions such as antibody-dependent cell-mediated cytotoxicity (ADCC) or complement-dependent cytotoxicity (CDC). Here, we provide a protocol for the generation of IgG-Fc de-galactosylated, galactosylated, de-sialylated and sialylated IgG antibodies using recombinant glycosidases and glycosyltransferases.
Keywords: Antibody glycosylationBackground
The use of glycosyltransferases for antibody glycan modification allows the attachment of sugar substrates to pre-existing glycan residues. Immunoglobulin G carries a single, highly conserved N-glycosylation site in each of its CH2 domains (Arnold et al., 2007) (Figure 1) allowing site-specific glycan modification with glycosyltransferases. Antibodies may carry additional N-glycans if their Fab domains contain Asn-X-Ser/Thr (X ≠ Pro) sequences (Mellquist et al., 1998). Careful selection of a monoclonal antibody lacking Fab glycosylation is therefore important for Fc-specific glycan modification. The protocol described herein was developed based on the following publications (Kingston, 2003; Kaneko et al., 2006; Anthony et al., 2008; Barb et al., 2009; Quast et al., 2015).

Figure 1. The IgG-Fc N-glycan. Schematic depiction of IgG with two fully processed IgG-Fc N-glycans (left) and composition of the glycan (right).
Materials and Reagents
- Amicon Ultra-4 Centrifugal Filter Units with 50 kDa MWCO (Merck Millipore Corporation, catalog number: UFC805096 )
- HisTrap HP column (GE Healthcare, catalog number: 29-0510-21 )
- StrepTrapTM HP column (GE Healthcare, catalog number: 29-0486-53 )
- Empty SPE tubes, 12 ml (Sigma-Aldrich, catalog number: 57176 )
- VisidryTM Female Luer Plug (Sigma-Aldrich, catalog number: 57098 )
- Caps for 12 ml SPE Tubes (Sigma-Aldrich, catalog number: 52174-U )
- 15 cm cell culture dish
- Spectra/Por 3 dialysis membrane (MWCO 3.5 kD), 45 mm width (VWR International, catalog number: 734-0686 )
- 10 clamps for dialysis tubing: Spectra/Por Closures, 55 mm (Spectrum, catalog number: 132737 )
- 0.2 µm syringe filters (Filtropur S plus 0.2) (Sarstedt AG, catalog number: 83.1826.102 )
- 0.2 µm bottle-top vacuum filter systems (Vakuumfiltration 1000 “rapid” Filtermax) (TTP, catalog number: 99950 )
- HEK293, HEK293T or HKB11 cells
- MES sodium salt (AppliChem GmbH, catalog number: A3101 )
- MES monohydrate (AppliChem GmbH, catalog number: A4730 )
- HEPES (Sigma-Aldrich, catalog number: H3375 )
- MOPS (AppliChem GmbH, catalog number: A1076 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9451 )
- Potassium hydroxide (KOH) (Sigma-Aldrich, catalog number: 306568 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: 71380 )
- Sodium citrate dihydrate (Sigma-Aldrich, catalog number: W302600-K )
- Citric acid (Sigma-Aldrich, catalog number: 251275 )
- Calcium chloride dihydrate (CaCl2·2H2O) (Sigma-Aldrich, catalog number: C7902 )
- MnCl2 tetrahydrate (AppliChem GmbH, catalog number: A2087 )
- MgCl2 hexahydrate (AppliChem GmbH, catalog number: A4425 )
- Sodium phosphate dibasic (Na2HPO4) (Sigma-Aldrich, catalog number: S3264 )
- Sodium dihydrogen phosphate monohydrate (NaH2PO4) (AppliChem GmbH, catalog number: A1047 )
- Potassium phosphate monobasic (KH2PO4) (Sigma-Aldrich, catalog number: P0662 )
- Trizma® hydrochloride (Tris-HCl) (Sigma-Aldrich, catalog number: T5941-500 G )
- Glycine (Sigma-Aldrich, catalog number: G8898 )
- Imidazole (AppliChem GmbH, catalog number: A1073 )
- Tween 20 (Sigma-Aldrich, catalog number: P7949 )
- Glycerol (Sigma-Aldrich, catalog number: G5516 )
- Sodium azide (AppliChem GmbH, catalog number: A1430 )
- D(+)-Lactose monohydrate (AppliChem GmbH, catalog number: A0880 )
- 1 M Tris, pH 8.8 (AppliChem GmbH, catalog number: A4265 )
- d-Desthiobiotin (Sigma-Aldrich, catalog number: D1411-500 MG )
- Penicillin and streptomycin, liquid (Thermo Fischer Scientific, GibcoTM, catalog number: 15070-063 )
- UDP-galactose (Merck Millipore Corporation, Calbiochem®, catalog number: 670111-50 MG )
- CMP-sialic acid (Merck Millipore Corporation, Calbiochem®, catalog number: 233264-5 MG )
- StarGate® pCSG-IBA-144 Acceptor Vectors (IBA, catalog number: 5-5144-001 )
- StarGate® transfer reagent kit (IBA, catalog number: 5-1603-001 )
- Western Blocking Reagent, Solution (Sigma-Aldrich, catalog number: 11921673001 )
- Neuraminidase from Clostridium perfringes, recombinantly expressed in E. coli (New England Biolabs, catalog number: P0720S )
- β1-4-galactosidase from Streptococcus pneumoniae, recombinantly expressed in E. coli (Merck Millipore Corporation, Calbiochem®, catalog number: 345806-50 MIU )
- DMEM high glucose (4.5 g/L) (Thermo Fischer Scientific, GibcoTM, catalog number: 11965092 )
- Protein G sepharose for fast flow (GE healthcare, catalog number: 17-0618-01 )
- 20 µM polyethylene (PE) frits (Sigma-Aldrich, catalog number: 57181 )
- Biotinylated Lens Culinaris agglutinin (LCA) (Reactolab, catalog number: B-1045 )
- Biotinylated Erythrina Cristagalli lectin (ECL) (Reactolab, catalog number: B-1145 )
- Biotinylated Elderberry bark lectin (Sambucus nigra agglutinin, SNA) (Reactolab, catalog number: B-1305 )
- Agarose bound Sambucus Nigra Lectin (SNA), Vector Laboratories (Reactolab, catalog number: AL1303 )
- Streptavidin-HRP (Mabtech, catalog number: 3310-9 )
- MEM NEAA (non-essential amino acids) 100x (Thermo Fischer Scientific, GibcoTM, catalog number: 1140050 )
- Sodium pyruvate solution (100 mM) (Sigma-Aldrich, catalog number: S8636 )
- PierceTM ECL Western Blotting Substrate (Thermo Fischer Scientific, catalog number: 32106 )
- SuperSignalTM West Pico Chemiluminescent Substrate (Thermo Fischer Scientific catalog number: 34080 )
- General
- Tris-buffered saline (TBS) (see Recipes)
- Phosphate-buffered saline (PBS) (see Recipes)
- Tris-buffered saline (TBS) (see Recipes)
- For purification of recombinant glycosyltransferases
- 293, 293T and HKB11 cell culture medium (see Recipes)
- 293, 293T and HKB11 cell protein expression medium (see Recipes)
- 2x HEPES-buffered saline (HeBS) (see Recipes)
- 2.5 M CaCl2 (see Recipes)
- Strep-trap column elution buffer (see Recipes)
- His-trap elution buffer (see Recipes)
- His-trap wash buffer (see Recipes)
- His-trap supernatant dilution buffer (see Recipes)
- 293, 293T and HKB11 cell culture medium (see Recipes)
- For enzymatic galactosylation
- Galactosyltransferase buffer (see Recipes)
- Galactosyltransferase buffer (see Recipes)
- For enzymatic sialylation
- Sialyltransferase buffer (see Recipes)
- Sialyltransferase buffer (see Recipes)
- For enrichment of highly-sialylated antibodies using SNA-agarose
- SNA-agarose binding/wash buffer (see Recipes)
- SNA-agarose elution Buffer (see Recipes)
- SNA-agarose clearing Buffer (see Recipes)
- 0.08% NaN3 (see Recipes)
- SNA-agarose binding/wash buffer (see Recipes)
- For de-sialylation using recombinant neuraminidase
- Neuraminidase buffer (see Recipes)
- Neuraminidase buffer (see Recipes)
- For de-galactosylation using recombinant β1-4-galactosidase
- β1-4-galactosidase buffer (see Recipes)
- β1-4-galactosidase buffer (see Recipes)
- For purification of glyco-modified antibodies using gravity flow protein-G sepharose columns
- Protein G column elution buffer (see Recipes)
- Protein G eluate neutralization buffer (see Recipes)
- Protein G column elution buffer (see Recipes)
- For analysis of antibody glycosylation by lectin-blotting
- Lectin blot incubation buffer (see Recipes)
- Lectin blot wash buffer (see Recipes)
- Lectin blot incubation buffer (see Recipes)
Equipment
- Spectrophotometer (Thermo Scientific, NanoDrop, model: 1000 )
- Tabletop centrifuge capable of > 20,000 x g centrifugation and cooling to 4 °C (Eppendorf)
- GE ÄKTAprime plus (GE Healthcare, catalog number: 11-0013-13 )
- Mini-PROTEAN® Tetra vertical electrophoresis cell, 4-gel, for 1.0 mm thick handcast gels, with PowerPacTM Basic power supply (Bio-Rad, catalog number: 1658025FC )
- Fusion FX7 gel imaging system (Vilber, model: Fusion FX7 )
Procedure
- Purification of recombinant glycosyltransferases
To obtain pure, functional sialyl- and galactosyltransferases, the respective coding sequences are cloned into eukaryotic expression vectors. These vectors contain a BM40 signal sequence resulting in secretion of the recombinant protein as well as N- and C-terminal affinity tags which are used for purification of the glycosyltransferases from cell culture supernatants of transiently transfected eukaryotic cells.- Clone the extracellular domain of human ST6Gal1 (NCBI Reference Sequence: NP_003023.1; from amino acid sequence KEKKKGS until GFRTIHC) and bovine β1,4GalT (GenBank: AAI20416.1 from amino acid sequence GSNLTSA until VDIGTPS) into pCSG-IBA-144 expression vectors according to the manufacturer’s recommendations. This results in a fusion protein containing an N-terminal Twin-Strep®-tag and a C-terminal 6xHistidine-tag.
- Transiently transfect HEK293, HEK293T or HKB11 cells with calcium-phosphate precipitated plasmids [adapted from (Kingston, 2003)]:
- Plate 6 x 106 cells per 15 cm cell culture dish in 20 ml DMEM high glucose containing P/S and 10% FCS.
- On the next day, freshly precipitate the plasmid DNA (values are given for one 15 cm cell culture dish and can be scaled up):
- Mix 20 µg plasmid DNA with 50 µl 2.5 M CaCl2 by pipetting up and down and fill up to 500 µl with ddH2O. Add 500 µl 2x HeBS in a 15 ml tube and slowly add the DNA-CaCl2 mix while creating bubbles using an electronic pipettor (Figure 2).
- Incubate for 30 min at room temperature.
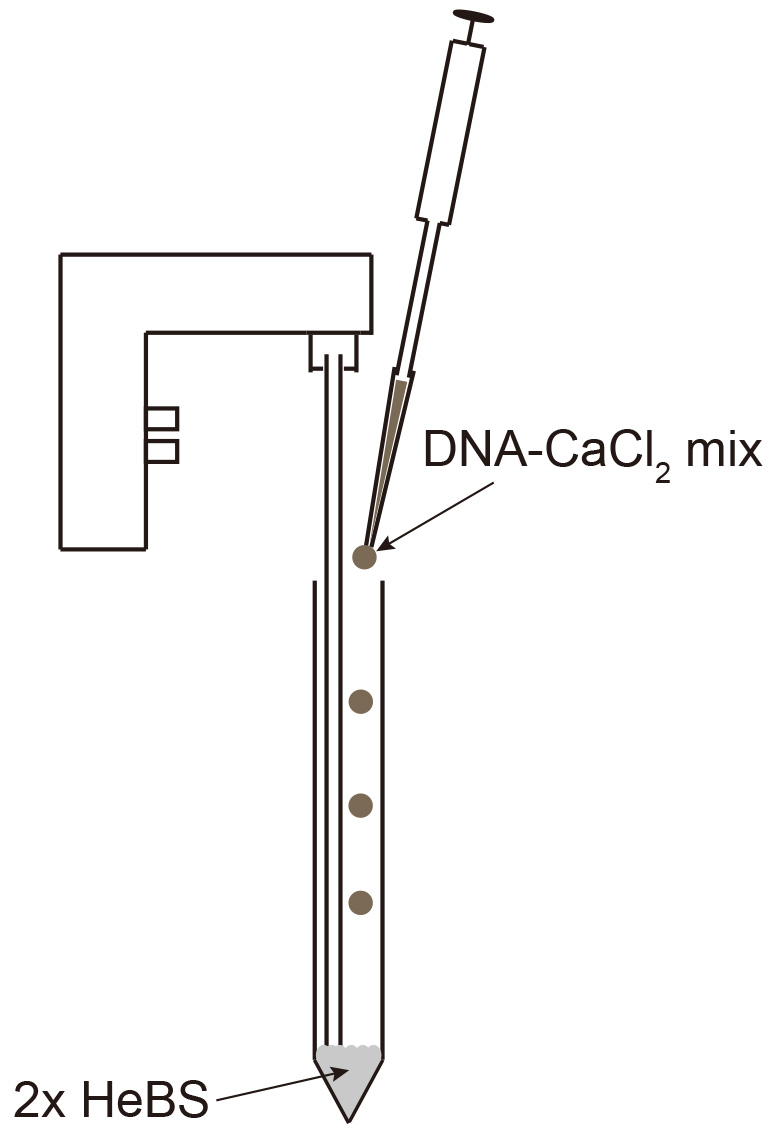
Figure 2. Mixing DNA-CaCl2 into 2x HeBS - Mix 20 µg plasmid DNA with 50 µl 2.5 M CaCl2 by pipetting up and down and fill up to 500 µl with ddH2O. Add 500 µl 2x HeBS in a 15 ml tube and slowly add the DNA-CaCl2 mix while creating bubbles using an electronic pipettor (Figure 2).
- Distribute equally over the plate seeded on the day before and mix by carefully tilting the plate.
- After 12-16 h, suck-off the medium and carefully replace it with 30 ml DMEM (4.5 g/L glucose) containing P/S, 10% FCS, NEAA and sodium pyruvate.
- Incubate for 4-5 days.
- Harvest the supernatant and sterile-filter through 0.2 µm.
- Immediately proceed to HisTrap and SterpTrap column mediated purification.
- Plate 6 x 106 cells per 15 cm cell culture dish in 20 ml DMEM high glucose containing P/S and 10% FCS.
- Purify the recombinant glycosyltransferases using HisTrap (nickel affinity) and StrepTrapTM (SII-tag affinity) columns
- Dilute the cell culture supernatant 1:2 with PBS containing 40 mM imidazole (pH 7.4).
- Apply to a 1 or 5 ml HIS-trap column (GE) using a GE ÄKTAprime plus.
- Wash with 10 column volumes PBS containing 20 mM imidazole.
- Elute with 10 column volumes 300 mM imidazole.
- Collect the entire eluate (the first 5-10 column volumes) and slowly (< 0.5 ml/min flow rate) apply to a StrepTrapTM column using GE ÄKTAprime plus.
- Wash with 10 column volumes PBS.
- Elute with 10 column volumes 2.5 mM d-desthiobiotin in PBS.
- Dialyze the protein-containing fractions by filling them into a Spectra/Por 3 Dialysis Membrane (MWCO 3.5 kD) tube and overnight incubation in at least 1 liter 0.2 M MES pH 6.5 (β1-4GalT) or 25 mM MOPS 100 mM KCl pH 7.2 (St6Gal).
- Determine the protein concentration using nano-drop. The absorption at 280 nm for a 1% (10 g/L) solution is 13.1 for β 1-4GalT and 17.1 for ST6Gal1. If necessary, concentrate using Amicon Ultra-4 Centrifugal Filter Units to reach a final concentration between 0.5 and 1.5 mg/ml.
- Sterilize by 0.2 µm filtration, add 20% sterile glycerol, aliquot and store at -20 °C.
- Dilute the cell culture supernatant 1:2 with PBS containing 40 mM imidazole (pH 7.4).
- Clone the extracellular domain of human ST6Gal1 (NCBI Reference Sequence: NP_003023.1; from amino acid sequence KEKKKGS until GFRTIHC) and bovine β1,4GalT (GenBank: AAI20416.1 from amino acid sequence GSNLTSA until VDIGTPS) into pCSG-IBA-144 expression vectors according to the manufacturer’s recommendations. This results in a fusion protein containing an N-terminal Twin-Strep®-tag and a C-terminal 6xHistidine-tag.
- Enzymatic galactosylation
Recombinant galactosyltransferase and UDP-galactose is used to add galactose to the IgG-Fc N-glycans of antibodies.- Buffer-exchange the antibody by dialysis to 0.2 M MES pH 6.5.
- Remove the antibody from dialysis and store the dialysis buffer at 4 °C for later use. Add 5 µg β 1-4GalT per mg antibody, add UDP-galactose to a final concentration of 10 mM, MnCl2 to a final concentration of 20 mM and supplement with 0.02% NaN3.
- Incubate for 48 h at 37 °C.
- Centrifuge for 2 h at 4 °C and > 20,000 x g to remove aggregates.
- Purify the galactosylated antibody using gravity-flow protein-G sepharose columns or continue to sialylation. A schematic depiction of galactosylated IgG is shown in Figure 3.
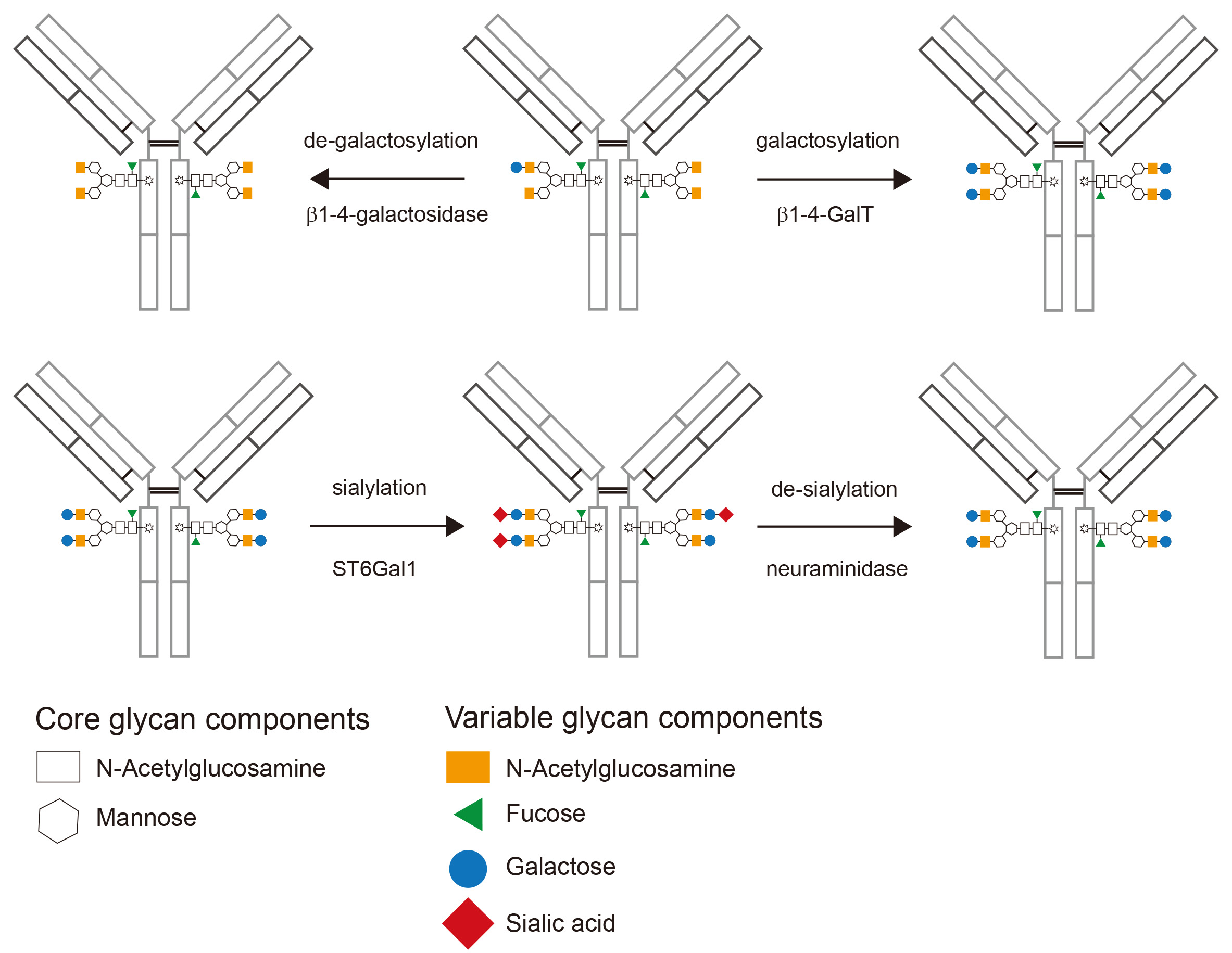
Figure 3. Schematic depiction of enzymatically glycomodified IgG. Starting from a typical glycan profile of a recombinantly expressed IgG (upper panel, center) the glycan composition after de-galactosylation (upper panel, left) and galactosylation (upper panel, right) is shown. After the addition of galactose, sialylation results in the addition of terminal sialic acid to most galactose residues (lower panel, center). Finally de-sialylation can be used to remove sialic acid (lower panel, right).
- Buffer-exchange the antibody by dialysis to 0.2 M MES pH 6.5.
- Enzymatic sialylation
Recombinant sialyltransferase and CMP-sialic acid is used to add sialic acid to galactose-containing IgG-Fc N-glycans.- Dialyze the entire galactosylation reaction overnight at 4 °C to 0.2 M MES, pH 6.5 using the same buffer as used for the initial dialysis in (B).
- Dialyze overnight at 4 °C to 25 mM MOPS, 100 mM KCl, pH 7.2.
- Remove the reaction from the dialysis and determine the protein concentration using Nano-Drop.
- Add 50 µg ST6Gal1 per mg antibody and CMP-sialic acid to a final concentration of 1.5 mM.
- Incubate for 24 h at 37 °C.
- Dialyze overnight at 4 °C to TBS.
- Purify the antibody using gravity-flow protein-G sepharose columns or enrich for highly sialylated antibodies using SNA-agarose. A schematic depiction of sialylated IgG is shown in Figure 3.
- Dialyze the entire galactosylation reaction overnight at 4 °C to 0.2 M MES, pH 6.5 using the same buffer as used for the initial dialysis in (B).
- Enrichment of highly-sialylated antibodies using SNA-agarose
The efficiency of enzymatic galactosylation and in particular sialylation can vary depending on the enzyme batch. The use of agarose bound Sambucus Nigra Lectin (SNA) which specifically binds to antibodies containing at least 2 sialic acid residues in the Fc-linked glycan region (Stadelmann et al., 2009) allows to enrich for antibodies with high sialic acid content.- Pour 1-2 ml SNA-agarose in an SPE tube containing a polyethylene (PE) frit with 20 µm porosity and let the slurry settle (this takes around 5 min). Place an additional PE frit on top of the slurry (Figure 4).
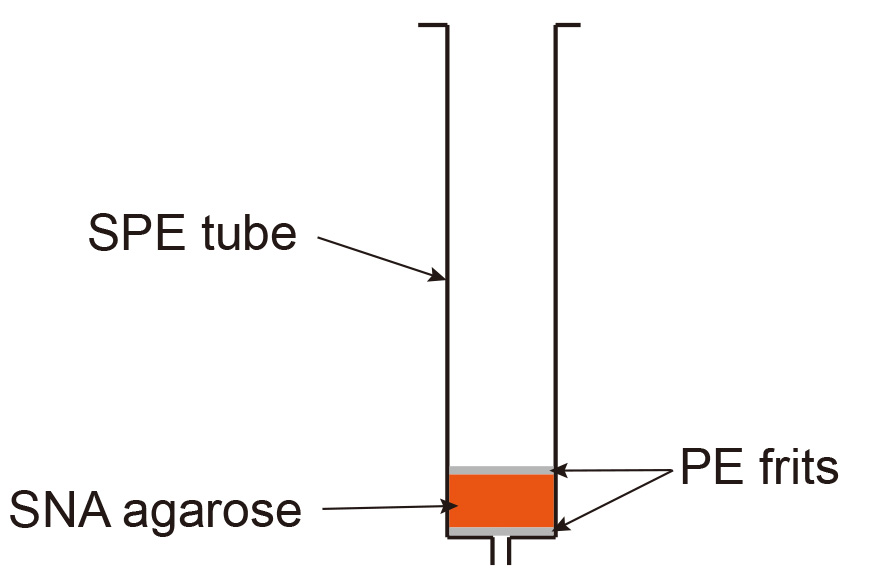
Figure 4. Set up of SNA-agarose gravity flow column
- Wash 3 times with 5 column volumes TBS + 0.1 M CaCl2.
- Add CaCl2 to a final concentration of 0.1 M to the sialylated antibodies previously dialyzed to TBS.
- Apply to the SNA-agarose column and collect the flow-through.
- Re-apply the flow through to the column.
- Wash 3 times with 5 column volumes TBS + 0.1 M CaCl2.
- Elute with 5 column volumes 0.5 M lactose in TBS (SNA agarose elution buffer) and collect the eluate in fractions corresponding to 1 column volume.
- Elute with 5 column volumes 0.5 M lactose/0.2 M acetic acid (SNA agarose clearing buffer) and collect the eluate in fractions corresponding to 1 column volume.
Regenerate the column by washing with 10 column volumes 0.5 M lactose/0.2 M acetic acid followed by 10 column volumes TBS and 5 column volumes SNA agarose storage buffer. Close the bottom of the column with a female luer plug, add 2 ml SNA agarose storage buffer and close the top with an SPE tube Cap. Store the column at 4 °C. - Dialyze the eluates to TBS.
- Purify using a protein-G sepharose column.
- Pour 1-2 ml SNA-agarose in an SPE tube containing a polyethylene (PE) frit with 20 µm porosity and let the slurry settle (this takes around 5 min). Place an additional PE frit on top of the slurry (Figure 4).
- De-sialylation using recombinant neuraminidase
- Dialyze the antibody to 50 mM sodium citrate, pH 6.0 (neuraminidase buffer).
- Add 70 units neuraminidase per mg antibody (or at least 300 U per ml).
- Incubate 48 h at 37 °C.
- Purify the antibody using protein-G sepharose. A schematic depiction of de-sialylated IgG is shown in Figure 3.
- Dialyze the antibody to 50 mM sodium citrate, pH 6.0 (neuraminidase buffer).
- De-galactosylation using recombinant β1-4-galactosidase
β-1-4-galactosidase can only remove terminal galactose (King et al., 2006). Therefore, neuraminidase treatment has to be done first if the antibody’s IgG-Fc glycan is sialylated.- Dialyze the antibody to 50 mM sodium phosphate buffer pH 6.0 (galactosidase buffer).
- Add 6 mU β1-4-galactosidase per 100 µg antibody.
- Incubate for 6 h at room temperature followed by 1 h at 37 °C.
- Purify the antibody using Protein-G sepharose. A schematic depiction of de-galactosylated IgG is shown in Figure 3.
- Dialyze the antibody to 50 mM sodium phosphate buffer pH 6.0 (galactosidase buffer).
- Purification of glyco-modified antibodies using gravity flow protein-G sepharose columns
This step is used to remove the glycosidases and/or glycosyltransferases added during glycol-modification procedures. Pour protein-G sepharose slurry (1-2 ml, depending on the amount of antibody to be purified) in an SPE tube containing a PE frit with 20 µm porosity. After the slurry has settled (this takes around 5 min), insert an additional SPE frit on top. The column is set up as shown for SNA agarose in Figure 4.- Apply the glycosylated antibody by gravity flow.
- Wash extensively with TBS.
- Elute with 0.1 M glycine, pH 2.5 and collect the eluates in 1 ml tubes containing the appropriate amount of 1 M Tris, pH 8.8 to result in a pH of 7.4.
- Dialyze to TBS or PBS.
- Apply the glycosylated antibody by gravity flow.
- Analysis of antibody glycosylation by lectin-blotting
Immunoblotting with the use of recombinant biotinylated lectins (referred to as lectin-blotting) can be used to detect the presence of specific glycan residues. This is a fast and reliable method to confirm the glycol-modification of antibodies.- Load 1 to 2 µg of each glycovariant on a 10% poly-acrylamide gel and transfer the protein to a 0.45 µm nitrocellulose membrane.
Note: Throughout the lectin-blotting procedure, avoid contact of the protein-containing side of the membrane with other surfaces, as this would lead to increased background signals in particular for the detection of sialic acid using biotinylated SNA. Therefore, use individual trays for each membrane throughout the procedure. - Block the membrane with Western Blocking Reagent (10% purified casein protein in maleic-acid buffer) diluted 1:10 in TBS at room temperature for 1 h or at 4 °C overnight.
- Remove the blocking solution and add the respective lectin for the glycan of interest (ECL for galactose, LCA for mannose and SNA for sialic acid) diluted 1:1,000 (resulting in 5 µg/ml for ECL, 5 µg/ml for LCA and 2 µg/ml for SNA) in TBS containing 1 mM CaCl2, MgCl2, MnCl2 and 1% Western Blocking Reagent (lectin incubation buffer).
- Incubate at room temperature for 1 h.
- Wash at least 6 x 5 min with TBS containing 0.1% Tween 20.
- Incubate with TBS containing 1% Western Blocking Reagent and Streptavidin-HRP (1:1,000) at room temperature for 1 h.
- Wash at least 6 x 5 min with TBS containing 0.1% Tween 20.
- Apply a chemiluminescent peroxide substrate and detect the signal using a gel imaging system or x-ray film. LCA and SNA work well using PierceTM ECL as substrate, ECL using SuperSignalTM West Pico (Figure 5).
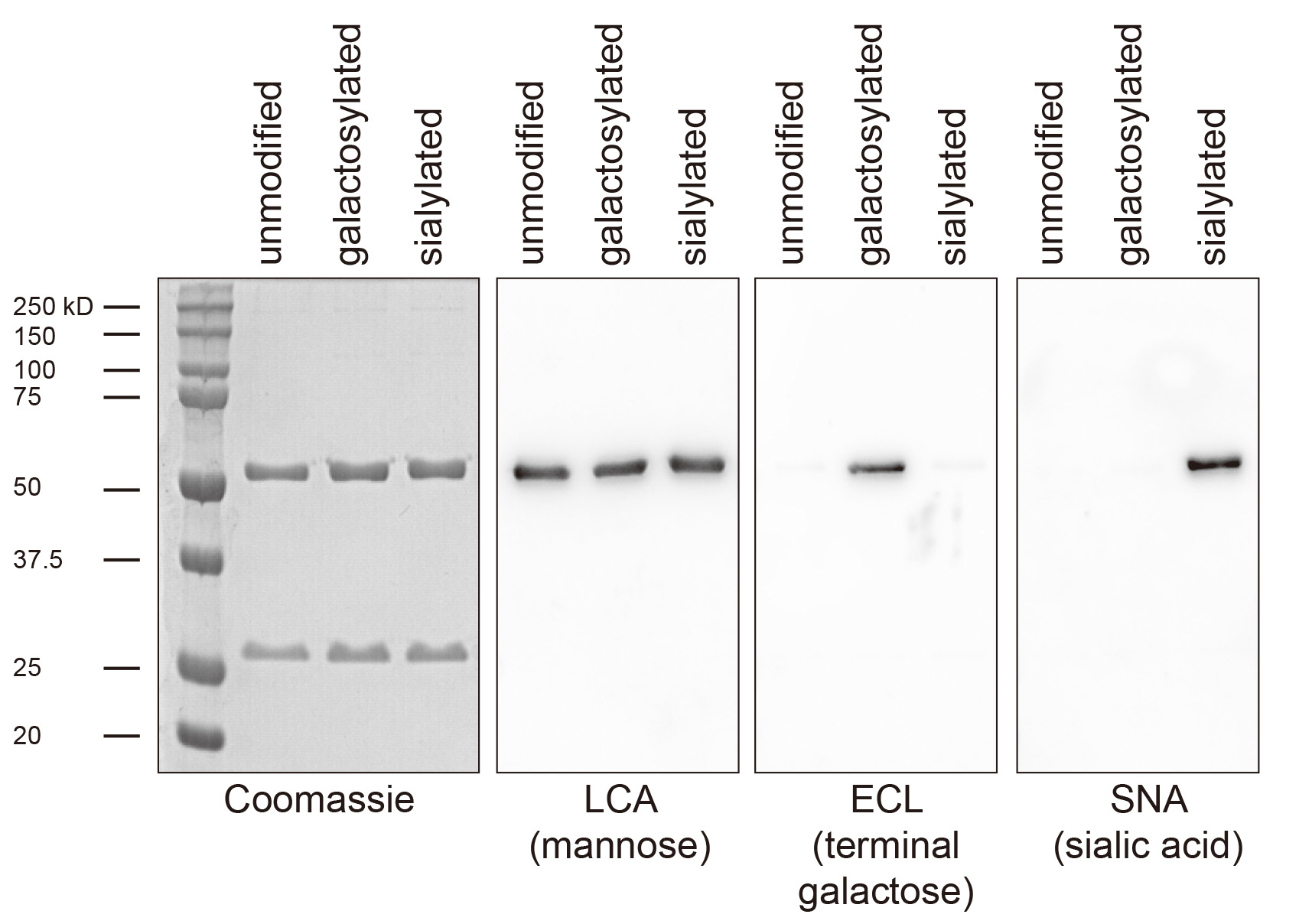
Figure 5. Exemplary lectin-blot analysis of antibody glycosylation. Shown is a 293T cell expressed Fc-humanized IgG1 antibody before glycosylation (unmodified), after galactosylation and purification using protein G sepharose (galactosylated) and after galactosylation, sialylation and purification using protein G sepharose (sialylated). Coomassie staining and immunoblotting for the core glycan component mannose is used to confirm purity and equal loading, respectively. ECL detects only terminal galactose and the signal is therefore lost upon sialylation.
- Load 1 to 2 µg of each glycovariant on a 10% poly-acrylamide gel and transfer the protein to a 0.45 µm nitrocellulose membrane.
Recipes
- General
- Tris-buffered saline (TBS)
0.05 M Tris-HCl
0.15 M NaCl
Adjust pH to 7.4 - Phosphate-buffered saline (PBS)
2.7 mM KCl
8 mM Na2HPO4
1.8 mM KH2PO4
137 mM NaCl
Adjust pH to 7.4
- Tris-buffered saline (TBS)
- For purification of recombinant glycosyltransferases
- 293, 293T and HKB11 cell culture medium
DMEM high glucose
Penicillin/streptomycin (P/S, 50 U/ml)
1 mM sodium pyruvate
10% fetal calf serum - 293, 293T and HKB11 cell protein expression medium
DMEM high glucose
Penicillin/streptomycin (P/S, 50 U/ml)
1x NEAA
1 mM sodium pyruvate
10% fetal calf serum - 2x HEPES-buffered saline (HeBS)
0.28 M CaCl2
0.05 M HEPES (N-2-hydroxyethylpiperazine-N’-2-ethanesulfonic acid)
1.5 mM Na2PO4
Adjust pH to 7.05 with 5 M NaOH and sterilize by 0.2 µm filtration - 2.5 M CaCl2
2.5 M CaCl2 in ddH2O - StrepTrapTM column elution buffer
2.5 mM D-desthiobiotin in PBS - His-trap elution buffer
300 mM imidazole in PBS, adjust pH to 7.4 - His-trap wash buffer
20 mM imidazole in PBS, adjust pH to 7.4 - His-trap supernatant dilution buffer
40 mM imidazole in PBS, adjust pH to 7.4
- 293, 293T and HKB11 cell culture medium
- For enzymatic galactosylation
- Galactosyltransferase buffer
25 mM MOPS
100 mM KCl
Adjust pH to pH 7.2 with KOH
- Galactosyltransferase buffer
- For enzymatic sialylation
- Sialyltransferase buffer
0.2 M MES
Prepare a 0.2 M MES-Sodium salt solution and a 0.2 M MES monohydrate solution and mix the two components to reach pH 6.5 (this requires around two fold excess of MES-sodium salt solution)
- Sialyltransferase buffer
- For enrichment of highly-sialylated antibodies using SNA-agarose
- SNA-agarose binding/wash buffer
TBS + 0.1 M CaCl2 - SNA-agarose elution Buffer
0.5 M lactose in TBS
Adjust pH to 7.5 - SNA-agarose clearing Buffer
0.5 M lactose in 0.2 M acetic acid - SNA-agarose storage Buffer
10 mM HEPES
0.15 M NaCl
0.1 mM CaCl2
0.08 % NaN3
20 mM lactose
Adjust pH to 7.5 with NaOH
- SNA-agarose binding/wash buffer
- For de-sialylation using recombinant neuraminidase
- Neuraminidase buffer
50 mM Sodium citrate, pH 6.0
Adjust pH to 6 with 50 mM citric acid
- Neuraminidase buffer
- For de-galactosylation using recombinant β1-4-galactosidase
- β1-4-galactosidase buffer
50 mM sodium phosphate, pH 6.0
Prepare a 50 mM Na2HPO4 solution and a 50 mM NaH2PO4 solution and mix the two components to reach pH 6.5 (this requires around 7 fold excess of 50 mM NaH2PO4)
- β1-4-galactosidase buffer
- For purification of glyco-modified antibodies using gravity flow protein-G sepharose columns
- Protein G column elution buffer
0.2 M Glycine
Adjust pH to 2.5 with HCl - Protein G eluate neutralization buffer
1 M Tris, pH 8.8
- Protein G column elution buffer
- For analysis of antibody glycosylation by lectin-blotting
- Lectin blot incubation buffer
TBS containing 1 mM CaCl2, 1 mM MnCl2, 1 mM MgCl2 and 1% Western Blocking Reagent - Lectin blot wash buffer
TBS + 0.1% Tween 20
- Lectin blot incubation buffer
Acknowledgments
We thank Dr. Falk Nimmerjahn (Department of Biology, University of Erlangen-Nuremberg, Erlangen, Germany) for his help in establishing this protocol and Patrick Weber (Institute of Experimental Immunology, Laboratory of Neuroinflammation, University of Zürich) for expert technical assistance. I. Quast was supported by a DOC scholarship provided by the Austrian Academy of Sciences (ÖAW). This protocol was adapted from Kingston, 2003; Kaneko et al., 2006; Anthony et al., 2008; Barb et al., 2009; Quast et al., 2015.
References
- Arnold, J. N., Wormald, M. R., Sim, R. B., Rudd, P. M. and Dwek, R. A. (2007). The impact of glycosylation on the biological function and structure of human immunoglobulins. Annu Rev Immunol 25: 21-50.
- Anthony, R. M., Nimmerjahn, F., Ashline, D. J., Reinhold, V. N., Paulson, J. C. and Ravetch, J. V. (2008). Recapitulation of IVIG anti-inflammatory activity with a recombinant IgG fc. Science 320(5874): 373-376.
- Barb, A. W., Brady, E. K. and Prestegard, J. H. (2009). Branch-specific sialylation of IgG-Fc glycans by ST6Gal-I. Biochemistry 48(41): 9705-9707.
- Kaneko, Y., Nimmerjahn, F. and Ravetch, E. V. (2006). Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation. Science 313(5787): 670-673.
- King, S. J., Hippe, K. R. and Weiser, J. N. (2006). Deglycosylation of human glycoconjugates by the sequential activities of exoglycosidases expressed by Streptococcus pneumoniae. Molecular Microbiology 59(3): 961-974.
- Kingston, R. E., (2003). Introduction of DNA into mammalian cells. Current Protocols in Molecular Biology, Chapter 9 (DOI: 10.1002/0471142727.mb0900s64).
- Mellquist, J. L., Kasturi, L., Spitalnik, S. L. and Shakin-Eshleman, S. H. (1998). The amino acid following an Asn-X-Ser/Thr sequon is an important determinant of N-linked core glycosylation efficiency. Biochemistry 37(19): 6833-6837.
- Quast, I., Keller, C. W., Maurer, M. A., Giddens, J. P., Tackenberg, B., Wang, L. X., Munz, C., Nimmerjahn, F., Dalakas, M. C. and Lunemann, J. D. (2015). Sialylation of IgG Fc domain impairs complement-dependent cytotoxicity. Journal of Clinical Investigation 125(11): 4160-4170.
- Stadlmann, J., Weber, A., Pabst, M., Anderle, H., Kunert, R., Ehrlich, H. J., Peter Schwarz, H. and Altmann, F. (2009). A close look at human IgG sialylation and subclass distribution after lectin fractionation. Proteomics 9(17): 4143-4153.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Quast, I., Maurer, M. A. and Lünemann, J. D. (2016). Generation of IgG-Fc Glycovariants Using Recombinant Glycosidases and Glycosyltransferases. Bio-protocol 6(15): e1886. DOI: 10.21769/BioProtoc.1886.
Category
Molecular Biology > Protein > Protein-protein interaction
Immunology > Antibody analysis > Antibody modification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









