- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantification of Chitinase Activity in Fusarium oxysporum
Published: Vol 6, Iss 15, Aug 5, 2016 DOI: 10.21769/BioProtoc.1882 Views: 13796
Reviewed by: Zhaohui LiuClaudia CatalanottiAndreea Daraba
Abstract
Fungal morphogenetic development requires modification and plasticity of the cell wall, which implies synthesis and remodelling of its components, including chitin and glucan. Thus chitinase and glucanase activities are crucial for cell-wall biogenesis and cell division. Quantification of chitinase activity might be useful to identify structural defects that could negatively influence growth and morphogenesis of some filamentous fungi like Fusarium oxysporum, which produces both intracellular and secreted chitinases. The chitinolytic enzymes are categorized based on their enzymatic action on chitin substrates. Endochitinases are defined as the enzymes catalyzing the random cleavage at internal points in the chitin chain. Exochitinases catalyze the progressive release of acetylchitobiose or N-acetylglucosamine from the non-reducing end of chitin, and thus, are referred to as chitobiosidase and β-N-acetylglucosaminidase, respectively. Here we describe a simple method to easily purify chitinases in order to compare both endo- and exo-chitinase activity of different F. oxysporum strains. The protocol can be adapted to any fungal species.
Keywords: Chitinase activityMaterials and Reagents
- Monodur nylon filters 15 µm diameter (Filtravibracion S.L., catalog number: Nylon-15 )
- Sterile plastic funnels (80 mm diameter) (Tecnylab, catalog number: 45000150 )
- Eppendorf tubes
- Sterile spatula (Instrumentación Científica, S.L., catalog number: 2004NE8044 )
- Microtiter plates (Thermo Fisher Scientific, catalog number: 2205 )
- Dialysis membranes, cut-off 12 kDa (Sigma-Aldrich, catalog number: D9527-100FT )
- Fusarium oxysporum f.sp. lycopersici microconida suspensions from wild type (J. Tello, Universidad de Almeria, Spain), the Δcon7-1 and the cΔcon7-1 mutant strains (Ruiz-Roldan et al., 2015)
- Glycerol (Merck, catalog number: 104092 )
- Potato dextrose broth medium (PDB) (Scharlau, catalog number: 01483 )
- Chitin (Sigma-Aldrich, catalog number: C6137 )
- Bio-Rad protein assay Dye reagent concentrate (Bio-Rad Laboratories, catalog number: 500-0006 )
- Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: 10 735 078 001 )
- Polyethylenglycol 35,000 (Sigma-Aldrich, catalog number: 81310 )
- Na2HPO4 (Merck, catalog number: 1.06586.0500 )
- NaH2PO4 monohydrate (Merck, catalog number: 567549 )
- MgSO4·7H2O (Merck, catalog number: 1058865.5000 )
- KH2PO4 (Merck, catalog number: 104873.1000 )
- KCl (Merck, catalog number: 104933.0500 )
- NH4NO3 (Merck, catalog number: 1.01187.1000 )
- FeSO4 (Merck, catalog number: 103965.0500 )
- ZnSO4·7H2O (Merck, catalog number: 108883 )
- MnSO4 monohydrate (Merck, catalog number: 105941 )
- Glucose (Coger SAS, catalog number: 24379.363 )
- NaCl (Merck, catalog number: 1.06404.5000 )
- Fluorimetric Chitinase Assay Kit (Sigma-Aldrich, catalog number: CS1030 ). Contents:
- Assay Buffer (25 ml) (Sigma-Aldrich, catalog number: A8730 )
- 4-Methylumbelliferyl N-acetyl-β-D-glucosaminide (5 mg) (Sigma-Aldrich, catalog number: M2133 )
- 4-Methylumbelliferyl β-D-N,N’-diacetylchitobioside hydrate (5 mg) (Sigma-Aldrich, catalog number: M9763 )
- 4-Methylumbelliferyl β-D-N,N’,N’’-triacetylchitotriose (5 mg) (Sigma-Aldrich, catalog number: M5639 )
- Chitinase from Trichoderma viride (1 mg) (Sigma-Aldrich, catalog number: C6242 )
- 4-Methylumbelliferone Standard Solution, 50 mg/ml (1 ml) (Sigma-Aldrich, catalog number: M3570 )
- Sodium carbonate (2 g) (Sigma-Aldrich, catalog number: S2127 )
- Dimethyl sulfoxide (DMSO) (1 ml) (Sigma-Aldrich, catalog number: D8418 )
- Assay Buffer (25 ml) (Sigma-Aldrich, catalog number: A8730 )
- 0.05 M phosphate buffer (see Recipes)
- Synthetic medium (SM) (see Recipes)
- Phosphate-buffered saline (PBS) (see Recipes)
- Strate stock solutions (20 mg/ml) (see Recipes)
- Strate working solutions (see Recipes)
- Chitinase control enzyme (see Recipes)
- Stop solution (sodium carbonate solution) (see Recipes)
- 4-Methylumbelliferone standard solution (see Recipes)
Equipment
- Hemocytometer (Thoma) (Marienfeld, catalog number: 06 407 10 )
- Mini-BeadBeater -16 homogenizer (BioSpec Products)
- Mortar and pestle
- Orbital incubator with shaking (Infors, model: Multitron Pro )
- Microcentrifuge (Eppendorf MiniSpin plus, Fisher Scientific)
- Fluorimeter (SpectraFluorPlus, TECAN, catalog number: F129005 )
- Water bath (Selecta, catalog number: 6000138 )
- Vortex (IKA, model: MS2 Minishaker )
Procedure
- Sample preparation
- F. oxysporum f.sp. lycopersici strains were stored at -80 °C with 30% glycerol as microconidial suspension.
- For fresh microconidia production, aliquots of frozen microconidial stocks were inoculated into 100 ml of potato dextrose broth (PDB) and incubated for 3 days at 150 rpm and 28 °C in an orbital incubator. Cultures were then filtered through Monodur nylon filters placed on funnels to separate mycelia (Figure 1A), and centrifuged at 3,020 x g for 5 min to collect microconidia. Finally, pellets containing fresh microconidia were resuspended into 1 ml sterile dH2O and counted using a hemocytometer.
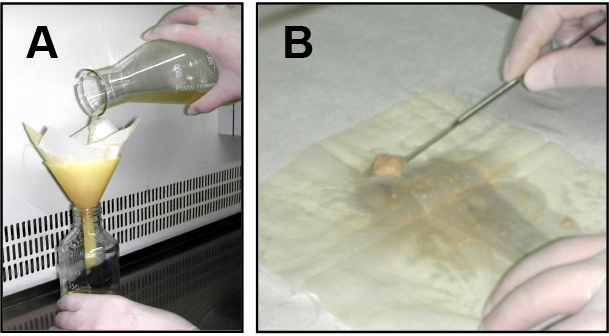
Figure 1. Filtration and mycelia harvesting procedure. A. Filtration of a fungal culture through a Monodur nylon membrane placed on a funnel. B. Subsequent harvesting of mycelia by scraping a nylon membrane using a spatula. - Aliquots containing 4 x 108 fresh microconidia from each strain were inoculated into 100 ml PDB and incubated in an orbital incubator at 28 °C with shaking at 170 rpm for 14 h (wild type and cΔcon7-1 strains) or 24 h (Δcon7-1 mutant).
Note: Conidial germination of Δcon7-1 mutant is significantly delayed compared to the wild type or the cΔcon7-1 strains. To normalize the growth delay of the mutant a longer incubation period was used for this strain. - Germlings were harvested by filtration through Monodur nylon membranes (Figure 1A), washed twice with sterile distilled water, scraped from the nylon filter using a spatula (Figure 1B) and directly transferred to 100 ml synthetic medium (SM) containing 0.5% chitin as sole carbon source.
- After 24 h growth, culture supernatants were filtered (to separate mycelia) and centrifuged (to discard spores) (as described in step A2). Supernatants were used to determine secreted chitinase activity, while mycelia were used for intracellular chitinase activity measurement.
- Culture supernatants were introduced into dialysis membranes that were sealed at both ends using plastic clamps, and subsequently submerged into 5 L distilled water for dialysis for 14 h at 4 °C.
Note: The dialysis step is necessary to eliminate remaining salts and nutrients from the culture medium. - After dialysis, membranes were transferred to a plastic tray, covered with PEG 35,000 powder and incubated at 4 °C until they were concentrated 100-fold (Figure 2). Samples were stored at -20 °C until use.

Figure 2. Concentration of culture supernatants using PEG 35,000 - Mycelia from step A5 were washed twice with sterile distilled water, frozen at -80 °C and ground with a mortar and pestle under liquid nitrogen.
- Samples were transferred to Eppendorf tubes containing 1ml phosphate buffer (0.05 M, pH 6.0) and homogenized by vortexing, followed by 2 min incubation at 4 °C.
- The homogenates were centrifuged 30 min at 10,000 x g and 4 °C, and the supernatants were collected and stored at -20 °C.
- F. oxysporum f.sp. lycopersici strains were stored at -80 °C with 30% glycerol as microconidial suspension.
- Quantification of protein content
The protein content of culture supernatants and mycelia was quantified according to the method of Bradford (Bradford, 1976) using the Bio-Rad protein assay kit with bovine serum albumin as standard.- Standard curve
- Dilute Bradford protein assay reagent 1:4 into distilled water.
- Aliquot 200 µl of diluted Bradford protein assay reagent per well into a microtiter plate.
- Prepare solutions containing serial dilutions of standard BSA ranging from 0 to 1 µg/ml.
- Start reaction by adding 5 µl of each standard BSA solution. Make at least two replicates of each standard dilution.
- Determine Absorbance at 595 nm using a Fluorimeter.
- Generate a standard curve for a series of BSA concentrations.
- Dilute Bradford protein assay reagent 1:4 into distilled water.
- Sample analysis
- Follow the directions given in Standard Curve, substituting BSA standard solutions for 5 µl fungal experimental samples. Please note that the total volume of the experimental sample assays should be equal to that of the BSA standard assays.
- Calculate the amount of protein in the experimental samples from the standard curve.
- Follow the directions given in Standard Curve, substituting BSA standard solutions for 5 µl fungal experimental samples. Please note that the total volume of the experimental sample assays should be equal to that of the BSA standard assays.
- Standard curve
- Enzyme activity determination
Endo- and exochitinase activities were individually quantified by using the Fluorimetric Chitinase Assay Kit following the manufacturer’s instructions. The kit provides three different substrates for the detection of the various types of the chitinolytic activity:
4-Methylumbelliferyl β-D-N,N’ -diacetyl-chitobioside, substrate suitable for exochitinase activity detection (chitobiosidase activity)
4-Methylumbelliferyl N-acetyl-β-D-glucosaminide, substrate suitable for exochitinase activity detection (-N-acetylglucosaminidase activity)
4-Methylumbelliferyl β-D-N,N’,N’’-triacetylchitotriose - substrate suitable for endochitinase activity detection
Chitinase activity is expressed as µg of 4-methylumbelliferone released from the substrate per µg of protein. Assays were performed in two independent experiments, each including two replicates.- Preparation of solutions (see Recipes 4-8)
- Assay
The chitinase hydrolysis is performed in an acidic environment (pH ~5.0) at 37 °C. The enzymatic hydrolysis liberates 4-methylumbelliferone (4MU). The fluorescence of liberated 4MU is measured in alkaline pH using a fluorimeter with excitation at 360 nm and emission at 450 nm. In order to quantitate the total chitinolytic activity, separate reactions should be run with the three substrates supplied in the kit. Note that in crude preparations there may be additive/synergist activity of different chitinases (i.e., 4-Methylumbelliferyl N,N’-diacetylchitobioside can be cleaved by β-N-acetylglucosaminidase and also chitobiosidase). It is recommended to perform the assays in duplicates.
For each substrate, perform a separate activity assay according to the following instructions of the manufacturer:- Equilibrate the substrate working solution(s) and the standard solution(s) to 37 °C by incubating for several minutes in a water bath.
- Set the fluorimeter at an excitation wavelength of 360 nm and an emission wavelength of 450 nm.
- Add the reaction components to 96 well plates according to Table 1 and mix by pipetting. Prepare the standard samples first (samples No. 4-8). Then, prepare the enzyme samples: add the substrate solution to the appropriate wells first and then add the enzyme (positive control or test sample).
Table 1.Reaction scheme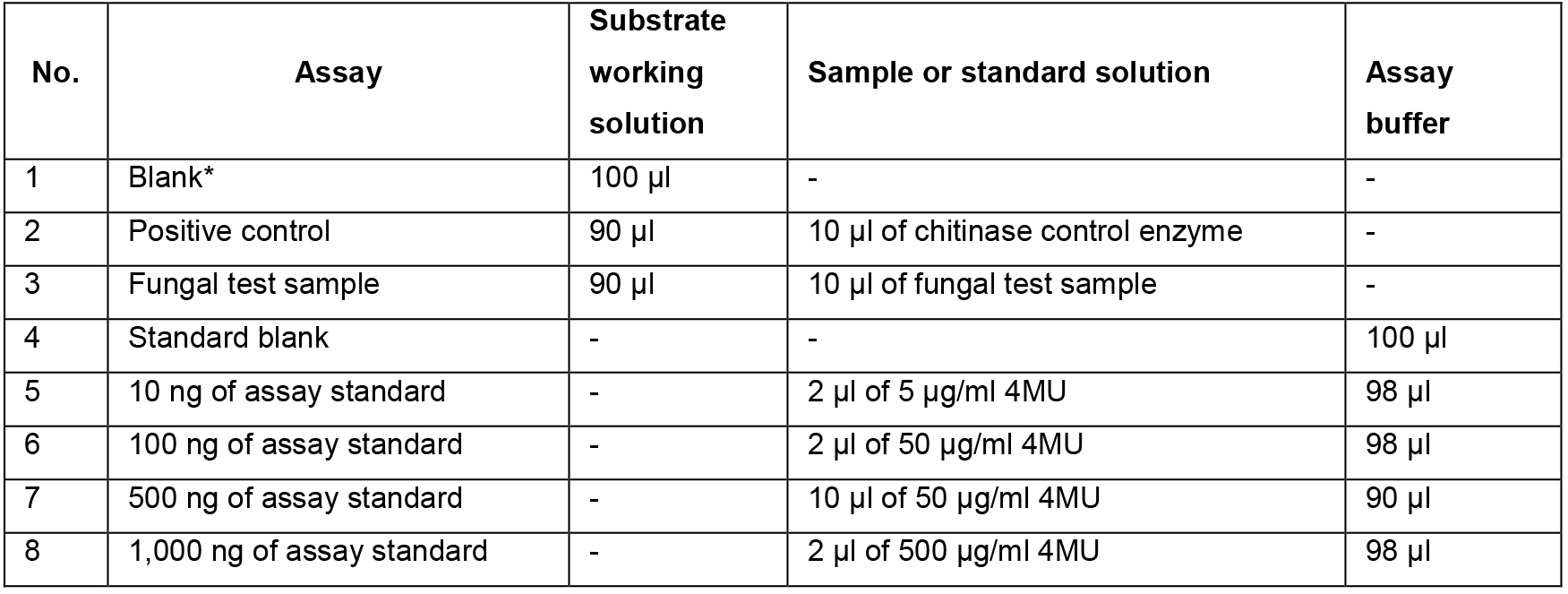
*A Blank reaction (substrate solution without enzyme) should be run to account for the spontaneous hydrolysis of the substrate during the incubation time. - Incubate the plate for 30 min at 37 °C. If required, the incubation time for highly active samples can be reduced to 15 min. On the other hand, in order to detect low levels of enzyme activity, the incubation time can be extended to 1 h.
- Stop the reactions by adding 200 µl of stop solution to each well.
- Measure the fluorescence at an excitation wavelength of 360 nm and an emission wavelength of 450 nm no later than 30 min after ending the reaction.
- Equilibrate the substrate working solution(s) and the standard solution(s) to 37 °C by incubating for several minutes in a water bath.
- Calculation
Unit definition: One unit of chitinase activity will release 1 mM of 4-methylumbelliferone from the appropriate substrate per minute at pH 5.0 at 37 °C.
The chitinase activity can be calculated using a standard curve prepared from the fluorescence readings of the 5 standard solutions (see Table 1).
Alternatively, the chitinase activity can be calculated using only a single standard concentration. It is recommended to measure the fluorescence of 100 ng (1.9 nmole/ml) Standard and then use the following equation: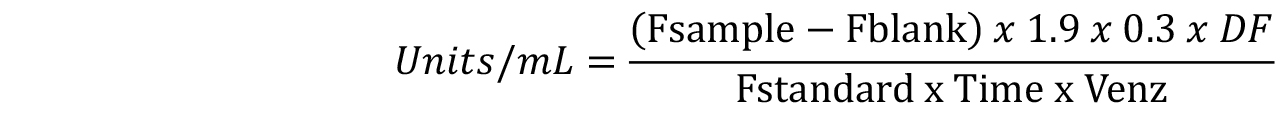
Where:
Fsample = fluorescence of the test sample
Fblank = fluorescence of the Blank (containing only substrate working solution)
0.3 = final reaction volume in milliliters after addition of the stop solution
DF = enzyme dilution factor
Fstandard = fluorescence of the standard solution minus the fluorescence of the Standard Blank
Time = min
Venz = volume of the sample in milliliter
Finally refer the resulting activity to the amount of protein calculated for each sample in previous steps.
- Preparation of solutions (see Recipes 4-8)
Representative data
Figure 3 shows a representative example of data obtained following this protocol.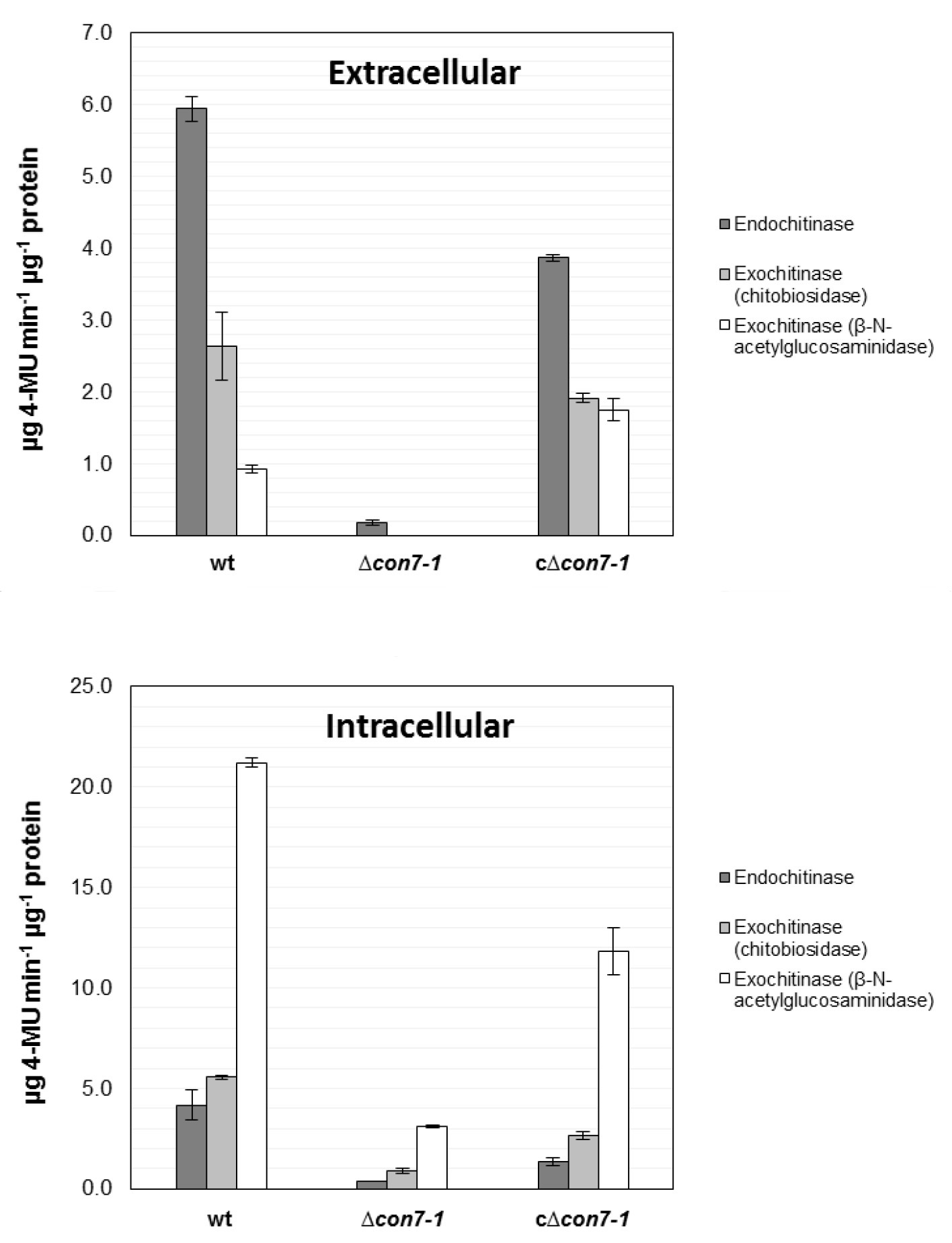
Figure 3. Quantification of intra- and extracellular chitinase activity. Intra- and extracellular protein extracts were obtained from germlings of the wild type strain (wt), ∆con7-1 mutant and the c∆con7-1 complemented strains after growth in SM supplemented with 0.5% chitin. Endo- and exochitinase (chitobiosidase and β-N-acetylglucosaminidase) activities were measured using the Fluorimetric Chitinase Assay Kit. Specific chitinase activity is expressed as µg of 4-methylumbelliferone (4MU) released from the appropriate substrate per µg of protein. Bars represent standard errors calculated from two independent experiments with two replicates each (Taken from Ruiz-Roldan et al., 2015).
Recipes
- 0.05 M phosphate buffer (pH 6.0)
Prepare the following solutions:- 1M Na2HPO4 (14.2 g in 100 ml distilled water)
- 1M NaH2PO4·H2O (13.8 g in 100 ml distilled water)
Dilute to 0.05 M final concentration - 1M Na2HPO4 (14.2 g in 100 ml distilled water)
- Synthetic medium (SM)
0.2 g/L MgSO4·7H2O
0.2 g/L KH2PO4
0.2 g/L KCl
1 g/L NH4NO3
0.01 g/L FeSO4
0.01 g/L ZnSO4·7H2O
0.01 g/L MnSO4.H2O
10 g/L glucose
Dissolve in deionized water and sterilize by autoclaving - Phosphate-buffered saline (PBS)
8 g/L NaCl
0.2 g/L KCl
1.44 g/L Na2HPO4
0.24 g/L KH2PO4
Dissolve the reagents listed above in 800 ml of H2O. Adjust the pH to 7.4 with HCl, and then add H2O to 1 L - Strate stock solutions (20 mg/ml)
- Add 0.25 ml of DMSO to the contents (5 mg) of each of the appropriate substrate bottle:
4-Methylumbelliferyl N-acetyl-β-D-glucosaminide, or
4-Methylumbelliferyl β-D-N,N’-diacetylchitobioside hydrate, or
4-Methylumbelliferyl β-D-N,N’,N’’-triacetylchitotriose - Vortex until dissolved
- Add 0.25 ml of DMSO to the contents (5 mg) of each of the appropriate substrate bottle:
- Strate working solutions
- Just before the assay dilute an aliquot of the substrate stock solution (20 mg/ml) 40-fold with Assay buffer to a concentration of 0.5 mg/ml.
- Mix by vortexing. Approximately 100 µl of substrate working solution are required for each test (each well).
- Just before the assay dilute an aliquot of the substrate stock solution (20 mg/ml) 40-fold with Assay buffer to a concentration of 0.5 mg/ml.
- Chitinase control enzyme
- Add 5 ml of PBS to the contents of the chitinase bottle to give a final chitinase concentration of 0.2 mg/ml.
- Vortex until dissolved. The chitinase dissolves immediately to give a slightly hazy solution. For long term storage, aliquots are stored at -20 °C (stable for at least 3 months at -20 °C).
- Just before use, dilute an aliquot of the chitinase 200-fold with PBS and use10 µl of the diluted enzyme per assay.
- Add 5 ml of PBS to the contents of the chitinase bottle to give a final chitinase concentration of 0.2 mg/ml.
- Stop solution (sodium carbonate solution)
- Add 47.2 ml of ultrapure water to the contents of the sodium carbonate bottle and mix well with a magnetic stirrer until completely dissolved.
- Store the stop solution at room temperature.
- Add 47.2 ml of ultrapure water to the contents of the sodium carbonate bottle and mix well with a magnetic stirrer until completely dissolved.
- 4-Methylumbelliferone standard solution
Before performing the assay, dilute an aliquot of the 4-Methylumbelliferone (4MU) standard solution in stop solution. Prepare 100-, 1,000-, and 10,000-fold dilutions to final concentrations of 500, 50 and 5 µg/ml, respectively. The final volume of the diluted standard solution in the assay should not exceed 10 µl.
Acknowledgments
This research was supported by the Spanish Ministerio de Economia y Competitividad (BIO2013-47870, and Ramon y Cajal Program), and Junta de Andalucia (P11-CVI-7319).
References
- Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248-254.
- Di Pietro, A. and Roncero, M. I. (1998). Cloning, expression, and role in pathogenicity of pg1 encoding the major extracellular endopolygalacturonase of the vascular wilt pathogen Fusarium oxysporum. Mol Plant Microbe Interact 11(2): 91-98.
- Pareja-Jaime, Y., Martin-Urdiroz, M., Roncero, M. I., Gonzalez-Reyes, J. A. and Roldan Mdel, C. (2010). Chitin synthase-deficient mutant of Fusarium oxysporum elicits tomato plant defence response and protects against wild-type infection. Mol Plant Pathol 11(4): 479-493.
- Ruiz-Roldan, C., Pareja-Jaime, Y., Gonzalez-Reyes, J. A. and MI, G. R. (2015). The transcription factor Con7-1 Is a master regulator of morphogenesis and virulence in Fusarium oxysporum. Mol Plant Microbe Interact 28(1): 55-68.
- Takaya, N., Yamazaki, D., Horiuchi, H., Ohta, A. and Takagi, M. (1998). Intracellular chitinase gene from Rhizopusoligosporus: molecular cloning and characterization. Microbiology 144 ( Pt 9): 2647-2654.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Ruiz-Roldan, C. and Roncero, M. I. G. (2016). Quantification of Chitinase Activity in Fusarium oxysporum. Bio-protocol 6(15): e1882. DOI: 10.21769/BioProtoc.1882.
Category
Biochemistry > Other compound > Chitinase
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link