- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Ubiquitination Assay for Mammalian Cells
Published: Vol 6, Iss 14, Jul 20, 2016 DOI: 10.21769/BioProtoc.1880 Views: 24538
Reviewed by: HongLok LungYu ChenAndrea Puhar

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Phos-tag Immunoblot Analysis for Detecting IRF5 Phosphorylation
Go R. Sato [...] Tomohiko Tamura
May 20, 2017 15417 Views

Novel Protein-oligonucleotide Conjugation Method Involving a High-affinity Capture HaloTag
Junshi Yazaki
Sep 20, 2020 5557 Views

Analysis of the Ubiquitination and Phosphorylation of Vangl Proteins
Di Feng [...] Bo Gao
Oct 20, 2022 3422 Views
Abstract
Ubiquitin is an 8.5 kDa protein that can be activated and conjugated by ubiquitin-activating enzyme E1 and ubiquitin-conjugating enzyme E2, respectively. Ubiquitin E3 ligases then recognize protein substrates, and then transfer the ubiquitin from E2 to the targeted protein. This biological process is called ubiquitination, and it is an important biological process which can signal protein degradation via the proteasome. The aim of this protocol is to describe a procedure that determines the level of cellular ubiquitination in a protein-of-interest relative to control cells.
Keywords: UbiquitinationMaterials and Reagents
- 10 cm dish
- Microcentrifuge tubes
- Eppendorf tubes
- Agarose beads
- U2OS cells
- HA-Ubiquitin plasmid
- FuGene 6 transfection reagent (Promega Corporation, FuGENE®, catalog number: E2691 )
- Reduced-serum medium (Thermo Fisher Scientific, Gibco®, catalog number: 31985-070 )
- MG132
- 1x PBS
- Trypsin (Thermo Fisher Scientific, Gibco®, catalog number: 25300 )
- IgG control antibody according to the targeted antibody species [e.g., Rabbit IgG (Santa Cruz Biotechnology, catalog number: sc-2027 )]
- Agarose conjugate [e.g., Protein A/G agarose (Santa Cruz Biotechnology, catalog number: sc-2003 )]
- BRCA1 antibody
- RIPA buffer (e.g., Santa Cruz Biotechnology, catalog number: sc-24948 )
- Anti-ubiquitin or anti-HA antibody
- Tris-HCl
- SDS
- Bromophenol blue
- β-mercaptoethanol
- Glycerol
- Loading buffer (see Recipes)
Equipment
- Tube rotator located in cold room 4 °C
- Centrifuges (Eppendorf, model: 5424/5424 R )
Procedure
- Co-transfect U2OS cells with HA-Ubiquitin plasmid with other plasmids of interest using FuGene 6. For a 10 cm dish, 3 μg of plasmid is mixed with 18 μl of Fugene 6 in 500 μl Reduced-serum medium, following the manufacturer's recommended procedure for use of Fugene 6. In the representative image, TUSC4 knockdown U2OS cells were used.
- After 15-30 min of incubation of the mixture at room temperature, add the mixture drop-wise onto the cell culture media of the 10 cm dish. Gently move the dish around to spread the mixture evenly throughout the dish.
- Harvest the cells within 48-72 h (10 µM MG132 incubation for 4-6 h prior to harvest is an optional step that enriches the ubiquitination signal).
- Prepare total cell lysate as described under a standard Western blot procedure. In short, wash the cells with 1x PBS and incubate cells with a volume of trypsin that is sufficient to detach the cells from the dish (e.g., 200 µl). Lyse the cells directly on the dish using RIPA buffer (e.g., 500 µl), then transfer the cells to a microcentrifuge tube.
- Disrupt the cells with gentle sonication (e.g., 3-sec pulse for 5 times).
- Centrifuge the sample at 10,000 x g for 10 min at 4 °C, then transfer the supernatant to a clean microcentrifuge tube on ice, without transferring the cellular debris at the bottom.
- (Optional step to reduce non-specific binding) Preclear cell lysate with appropriate IgG mouse control antibody (e.g., 0.25 µg antibody per 1 ml cell lysate), and 20 μl suspended agarose conjugate (e.g., Protein A/G agarose) and rotate mixture at 4 °C for 30 min to 1 h.
- Spin down the mixture at 1,000 x g for 1 min at 4 °C and transfer the supernantant containing the targeted protein into a clean microcentrifuge tube (a typical amount of protein is 2-5 mg). Incubate samples with an amount of primary antibody that is suggested by the manufacture at 4 °C with rotation for 2 h to overnight. In the representative image, BRCA1 antibody was used (Lu et al., 2012).
- Add 20 µl of appropriate agarose beads and incubate the samples at 4 °C with rotation for 1-2 h.
- Spin down the agarose beads at 1,000 x g for 1 min at 4 °C, discard the supernatant with care to not disturb the beads.
- Wash the beads with RIPA buffer or PBS 3 times with constant rotation at 4 °C, 15 min each time. RIPA buffer is a relatively strong detergent which can disrupt protein-protein interactions; alternate detergents may be considered for weaker protein-protein interactions. After the last wash, remove as much buffer as possible with minimal disruption of the beads.
Note: In some case, RIPA would be too harsh, so this needs to be adjusted case by case. - Add 20-50 µl of loading buffer and mix to re-suspend the beads. Incubate the samples for 5 min at 95 °C, and then subject to SDS-PAGE analysis or store the samples in -20 °C for future use. In the representative image, a 10% gel was used. Regular western blot condition is sufficient; some blots may require extended wash due to high background that can be caused by non-specific binding during the immunoprecipitation step.
- Blots can be probed using anti-ubiquitin or anti-HA antibody to determine the primary proteins’ ubiquitination levels.
Representative data
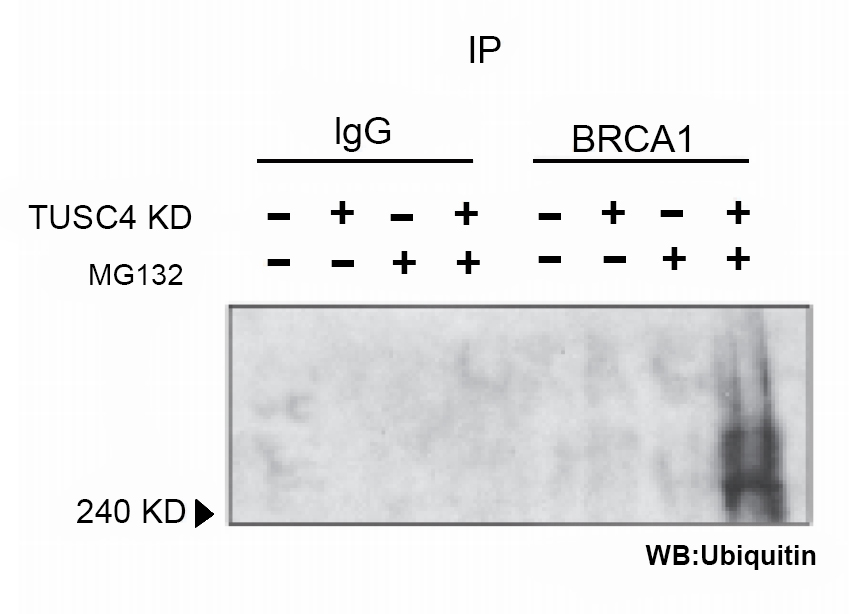
Figure 1. Knockdown of TUSC4 expression enhanced BRCA1 polyubiquitination, leading to BRCA1 protein degradation
NPRL2/TUSC4 knockdown increased ubiquitination level of BRCA1 compared with control cells (after MG132 enrichment for ubiquitination) (Peng et al., 2015).
Notes
- Preclear cell lysate with appropriate IgG control antibody is a necessary step to reduce background signals.
Recipes
- Loading buffer
62.5 mM Tris-HCl (pH 6.8)
2.5% SDS
0.002% bromophenol blue
0.7135 M (5%) β-mercaptoethanol
10% glycerol
Acknowledgments
This work was supported by The Department of Defense (DOD) Breast Cancer Research Program (BCRP) Era of Hope Scholar Award (W81XWH-10-1-0558; S.-Y. Lin).
References
- Lu, Y., Li, J., Cheng, D., Parameswaren, B., Zhang, S., Jiang, Z.,Yew, P. R., Peng, J., Ye, Q., Hu, Y. (2012). The F-box protein FBXO44 mediates BRCA1 ubiquitination and degradation. J Biol Chem 287:41014-22.
- Peng, Y., Dai, H., Wang, E., Lin, C.C., Mo, W., Peng, G., Lin, S.Y. (2015). TUSC4 Functions as a Tumor Suppressor by Regulating BRCA1 Stability. Cancer Res 75(2):378-86.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Peng, Y., Wang, E., Peng, G. and Lin, S. (2016). Ubiquitination Assay for Mammalian Cells . Bio-protocol 6(14): e1880. DOI: 10.21769/BioProtoc.1880.
Category
Biochemistry > Protein > Modification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









