- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Determination of Intracellular ATP Levels in Mycelium of Fusarium oxysporum
Published: Vol 6, Iss 14, Jul 20, 2016 DOI: 10.21769/BioProtoc.1869 Views: 10129
Reviewed by: Zhaohui LiuShahin S. AliSusheel KumarAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Intracellular Assessment of ATP Levels in Caenorhabditis elegans
Konstantinos Palikaras and Nektarios Tavernarakis
Dec 5, 2016 11578 Views

Measuring in vitro ATPase Activity with High Sensitivity Using Radiolabeled ATP
Sarina Veit and Thomas Günther Pomorski
May 20, 2023 2281 Views

RNA Cap Methyltransferase Activity Assay
Jackson B. Trotman and Daniel R. Schoenberg
Mar 20, 2018 8462 Views
Abstract
Glycolysis provides metabolites for energy production via oxidative phosphorylation during vegetative growth of Fusarium oxysporum. Therefore, determination of intracellular ATP levels might be of valuable help to analyze regulation of glycolysis/gluconeogenesis pathways. The protocol described here can be applied to other filamentous fungi.
Materials and Reagents
- Monodur nylon filters 15 µm diameter (Filtravibracion S.L., Spain, catalog number: Nylon-15 )
- Sterile plastic funnels (80 mm diameter) (Tecnylab, catalog number: 45000150 )
- Eppendorf tube (2 ml)
- 5 mm-diameter glass beads (Sigma-Aldrich, USA, catalog number: 18406-500G )
- Microtiter plates (TermoFisher Scientific, catalog number: 2205 )
- Fusarium oxysporum f.sp. lycopersici microconida suspensions from wild type and mutant strains.
Note: Strains used in this study are wild type strain 4287 (race 2) and Δcon7-1 and cΔcon7-1 mutant strains. - Glycerol (Merck, catalog number: 104092 )
- Potato dextrose broth (PDB) (Scharlau, Spain, catalog number: 01483 )
- Sterile dH2O
- Trichloroacetic acid (Sigma-Aldrich, catalog number: T6399 )
- EDTA (Amresco, catalog number: 0322 )
- MgSO4·7H2O (Merck, catalog number: 1058865.5000 )
- KH2PO4 (Merck, catalog number: 104873.1000 )
- KCl (Merck, catalog number: 104933.0500 )
- NH4NO3 (Merck, catalog number: 101187.1000 )
- FeSO4 (Merck, catalog number: 103965.0500 )
- ZnSO4·7H2O (Merck, catalog number: 108883 )
- MnSO4 monohydrate (Merck, catalog number: 105941 )
- Glucose (Coger SAS, catalog number: 24379.363 )
- ATP determination kit (Thermo Fischer Scientific, Molecular Probes®, catalog number: A22066 ).
Kit contents: - D-Luciferin (Component A, MW 302, blue cap), 5 vials, each containing 3 mg of lyophilized powder
- Luciferase, firefly recombinant (Component B, red cap) 40 μl of a 5 mg/ml solution in 25 mMTris-acetate, pH 7.8, 0.2 M ammonium sulfate, 15% (v/v) glycerol and 30% (v/v) ethylene glycol
- Dithiothreitol (DTT) (Component C, MW 154, black cap) 25 mg
- Adenosine 5’-triphosphate (ATP) (Component D, Green cap), 400 μl of a 5 mM solution in TE buffer
- 20x Reaction Buffer (Component E) 10 ml of 500 mM
- Tricine buffer, pH 7.8, 100 mM MgSO4, 2 mM EDTA and 2 mM sodium azide
- Extraction buffer (see Recipes)
- Potato dextrose broth (PDB) medium (see Recipes)
Equipment
- Sterile spatula (Fisher Scientific, catalog number: S50821 )
- Mini-BeadBeater-16 homogenizer (BioSpec Products)
- Hemocytometer Thoma (Marienfeld, Germany, catalog number: 06 407 10 )
- Orbital incubator (Infors, Multitron Pro, Switzerland)
- Microcentrifuge (Eppendorf, MiniSpin plus) (Fisher Scientific)
- Fluorimeter (TECAN, SpectraFluorPlus, catalog number: InfiniteM200PRO )
Procedure
- F. oxysporum f.sp. lycopersici wild type strain 4287 (race 2) was obtained from J. Tello, Universidad de Almeria, Spain. Δcon7-1 and cΔcon7-1 strains were described previously (Ruiz-Roldan et al., 20015). Strains are stored at -80 °C with 30 % glycerol as microconidial suspension.
- For fresh microconidia production, aliquots of frozen microconidial stocks are inoculated into 100 ml of potato dextrose broth (PDB) and incubated for 3 days at 150 rpm and 28 °C in an orbital incubator. Cultures are then filtered through Monodur nylon filters placed on funnels to separate mycelia (Figure 1A), and centrifuged at 3,020 x g for 5 min to collect microconidia. Finally, pellets containing fresh microconidia are resuspended into 1 ml sterile dH2O and counted using a hemocytometer.
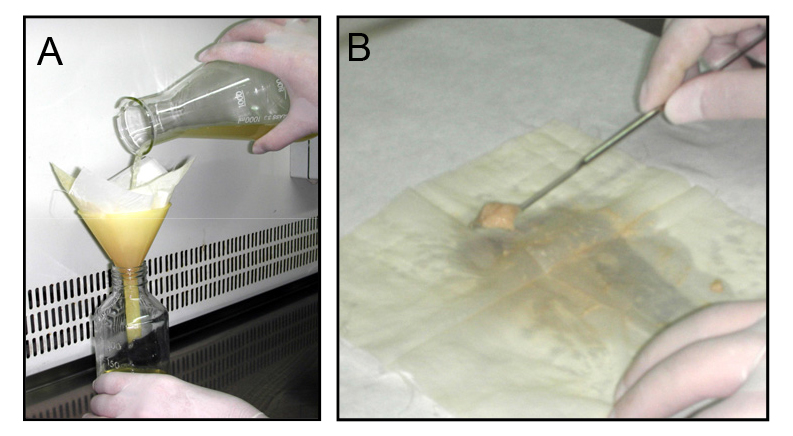
Figure 1. Filtration and mycelia harvesting procedure. A. Filtration of a fungal culture through a Monodur nylon membrane placed on a funnel. B. Subsequent harvesting of mycelia by scraping a nylon membrane using a spatula. - Aliquots containing 2.5 x 108 fresh microconidia are grown in 100 ml PDB at 170 rpm and 28 °C for 14 h (wild-type and cΔcon7-1 strains) or 24 h (Δcon7-1 mutant).
- After harvesting by Monodur filtration, mycelia are washed twice with sterile dH2O, separated from the filter by scraping using a spatula (Figure 1B) and transferred to 2 ml-Eppendorf tubes containing a 5 mm-diameter glass bead.
- Cells are resuspended into 1 ml of extraction buffer (380 mM trichloroacetic acid and 12.7 mM EDTA) and disrupted by 3 cycles of 30 sec agitation each using a Mini-BeadBeater homogenizer, followed by incubation at room temperature with shaking at 250 rpm for 15 min.
- The supernatant is harvested by centrifugation at 11,336 x g for 15 min at 4 °C. Aliquots (10 μl each) are used for quantification of ATP levels using the ATP determination kit (http://tools.thermofisher.com/content/sfs/manuals/mp22066.pdf) following these instructions of the manufacturer with some modifications (indicated in underlines):
- Reagent preparation
- Make 1.0 ml of 1x Reaction Buffer by adding 50 μl of 20x Reaction Buffer (Component E) to 950 μl of deionized water (dH2O). This volume will be sufficient to make 1 ml of 10 mM D-luciferin stock solution.
- Make 1 ml of a 10 mM D-luciferin stock solution by adding 1 ml of 1x Reaction Buffer (prepared in the previous step) to one vial of D-luciferin (Component A, blue cap). Protect from light until use. The D-luciferin stock solution is reasonably stable for several weeks if stored at ≤ -20 °C, protected from light.
- Prepare a 100 mM DTT stock solution by adding 1.62 ml of dH2O to the bottle containing 25 mg of DTT (Component C, black cap). Aliquot into ten 160 μl volumes and store frozen at ≤ -20 °C. Stock solutions of DTT stored properly are stable for six months to one year. Thawed aliquots are kept on ice or at 4°C until use.
- Prepare low-concentration ATP standard solutions by diluting the 5 mM ATP solution (Component D, green cap) in dH2O. The concentrations and volumes to make depend upon the sensitivity and the luminometer used. Typically, ATP concentrations ranging from 1 nM to 1 μM are appropriate. These dilute solutions are stable for several weeks when stored at ≤ -20°C.
- Standard reaction solution
- The manufacturer suggests combining the components of the reaction as follows to make 10 ml of a standard reaction solution. Adjust the volumes according to particular requirements.
8.9 ml dH2O
0.5 ml 20x Reaction Buffer (Component E)
0.1 ml 0.1 M DTT (from the previous step)
0.5 ml of 10 mM D-luciferin (from the previous step, store the remaining 0.5 ml at ≤ -20 °C for up to several weeks)
2.5 μl of firefly luciferase 5 mg/ml stock solution - Gently invert the tube to mix, do not vortex; the firefly luciferase enzyme is easily denatured. Keep the reaction solution protected from light until use, it can be stored at 2-6 °C protected from light for several days.
- Standard curve
- Place 190 µl aliquots of the standard reaction solution (prepared in previous steps) in a microtiter plate and measure the background luminescence using a flurorometer.
- Start the reaction by adding 10 µl of diluted ATP standard solutions (prepared in previous steps) and read the luminescence. The volume of the dilute ATP standard solution that is added to the standard assay solution (prepared in the previous step) should be less than 10% of the total assay volume. For example, a 100 μl total assay volume should contain 10 μl or less of the ATP standard solution.
- Subtract the background luminescence.
- Generate a standard curve for a series of ATP concentrations (Figure 2). Be sure to always add a constant sample volume of the ATP containing solution. R-values of around 0.9 are normally obtained with this kit.
- Sample analysis
- Follow the directions given in Standard Curve, substituting ATP standard solutions for 10 µl fungal experimental samples. Please note that the total volume of the experimental sample assays should be equal to that of the ATP standard assays, with the amount of sample added amounting to no more than 10% of the total assay volume.
- Calculate the amount of ATP in the experimental samples from the standard curve. The assay is repeated three times with independent biological samples.
Representative data
- Figure 2 shows a representative standard curve for calculation of ATP concentration.
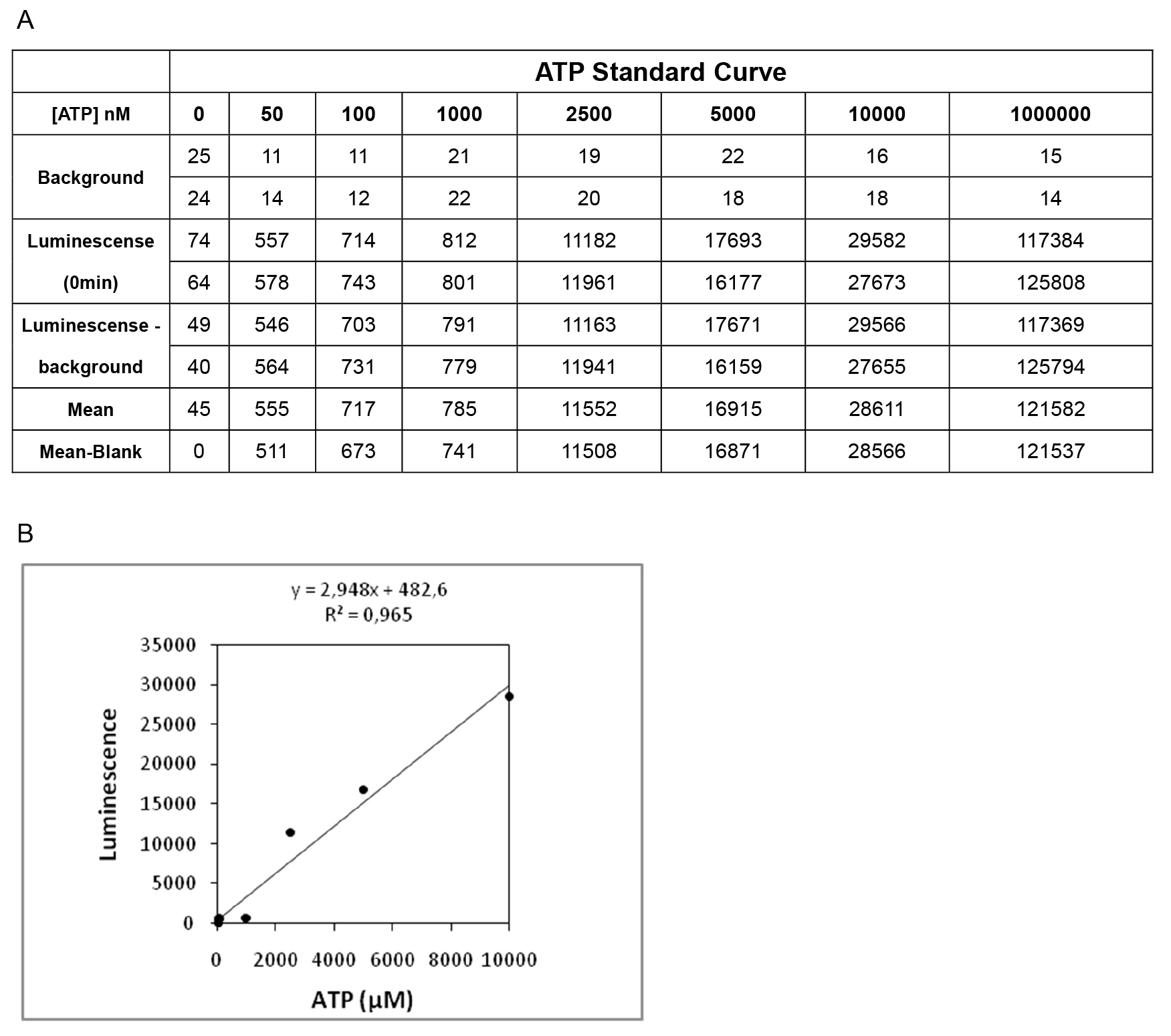
Figure 2. A representative standard curve for calculation of ATP concentration. A. Representative Luminescence values of the diluted ATP standard solutions. B. Graphical representation of Luminescence values (Y-axis) vs. nM of ATP of the standards (X-axis).
In this example, a luminescence value of 20,000 indicates that our test sample contains: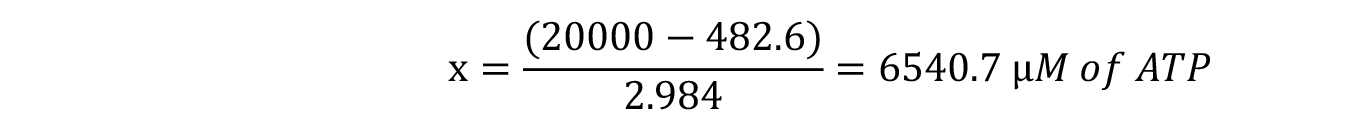
- Figure 3 shows a representative example of data obtained following this protocol.
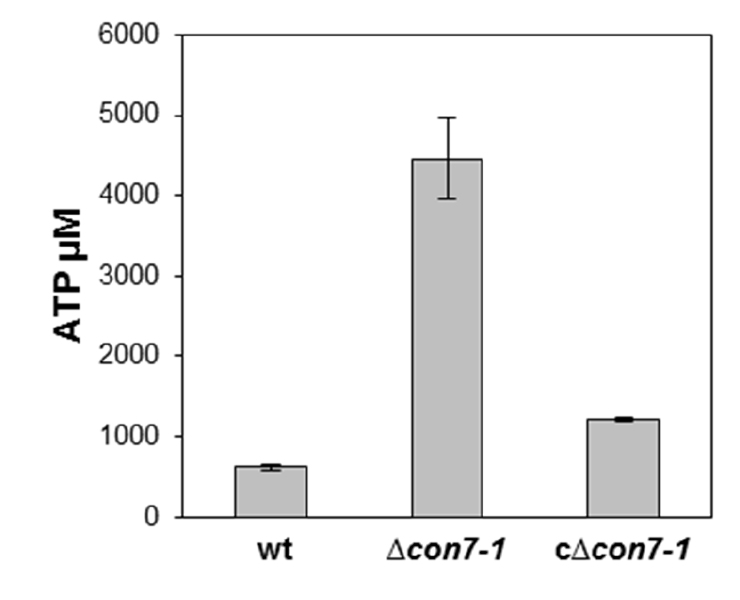 Figure 3. Intracellular ATP levels present in germlings of the indicated strains were quantified using the ATP determination kit (Molecular Probes)
Figure 3. Intracellular ATP levels present in germlings of the indicated strains were quantified using the ATP determination kit (Molecular Probes)
Recipes
- Extraction buffer
380 mM trichloroacetic acid
12.7 mM EDTA
Dissolve in deionized water - Potato dextrose broth medium (PDB)
Dissolve 24 g Potato dextrose broth in 1 L distilled water and sterilize by autoclaving
Acknowledgments
This research was supported by Junta de Andalucia (Proyecto de Excelencia CVI-7319) and the Spanish Ministerio de Economia y Competitividad (grant BIO2013-47870 and the Ramon y Cajal Program). This protocol was adapted from Ruiz-Roldan et al., 2015.
References
- Di Pietro, A. and Roncero, M. I. (1998). Cloning, expression, and role in pathogenicity of pg1 encoding the major extracellular endopolygalacturonase of the vascular wilt pathogen Fusarium oxysporum. Mol Plant Microbe Interact 11(2): 91-98.
- Herreros, E., Martinez, C. M., Almela, M. J., Marriott, M. S., De Las Heras, F. G. and Gargallo-Viola, D. (1998). Sordarins: in vitro activities of new antifungal derivatives against pathogenic yeasts, Pneumocystis carinii, and filamentous fungi. Antimicrob Agents Chemother 42(11): 2863-2869.
- Palicz, A., Foubert, T. R., Jesaitis, A. J., Marodi, L. and McPhail, L. C. (2001). Phosphatidic acid and diacylglycerol directly activate NADPH oxidase by interacting with enzyme components. J BiolChem 276(5): 3090-3097.
- Ruiz-Roldan, C., Pareja-Jaime, Y., Gonzalez-Reyes, J. A. and MI, G. R. (2015). The transcription factor Con7-1 is a master regulator of morphogenesis and virulence in Fusarium oxysporum. Mol Plant Microbe Interact 28(1): 55-68.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Ruiz-Roldan, C. and Roncero, M. I. G. (2016). Determination of Intracellular ATP Levels in Mycelium of Fusarium oxysporum. Bio-protocol 6(14): e1869. DOI: 10.21769/BioProtoc.1869.
Category
Biochemistry > Other compound > Nucleoside triphosphate > ATP
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link