- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Identification of Natural Hybrids by SSR Markers in Mussaenda
Published: Vol 6, Iss 13, Jul 5, 2016 DOI: 10.21769/BioProtoc.1853 Views: 10083
Reviewed by: Samik BhattacharyaGazala AmeenAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Experimental Pipeline for SNP and SSR Discovery and Genotyping Analysis of Mango (Mangifera indica L.)
Michal Sharabi-Schwager [...] Ron Ophir
Aug 20, 2016 11559 Views

Investigation of Transposon DNA Methylation and Copy Number Variation in Plants Using Southern Hybridisation
Vivek Hari Sundar G. and P. V. Shivaprasad
Jun 5, 2022 3411 Views

CAPS-Based SNP Genotyping for Nitrogen-Response Phenotypes in Maize Hybrids
Jannis Jacobs [...] Peter K. Lundquist
Dec 20, 2025 550 Views
Abstract
Detection of natural hybrids is of great significance for plant taxonomy, reproductive biology, and population genetic studies. Compared with methods depending on morphological characters, molecular markers provide reliable and much more accurate results. This protocol describes approaches employing microsatellite (SSR) markers to identify inter-specific hybrids in Mussaenda (Rubiaceae).
Keywords: Gene flowMaterials and Reagents
- Consumables
- Plant material
- Fresh leaves of the putative hybrid individuals, as well as the parental species, were collected and dried in silica gel. It usually takes 3-5 days for complete drying of the leaves.
- Fresh leaves of the putative hybrid individuals, as well as the parental species, were collected and dried in silica gel. It usually takes 3-5 days for complete drying of the leaves.
- Chemicals
- Taq PCR mastermix (Tiangen, catalog number: KT201 )
- Liquid nitrogen
- Absolute ethanol (Sinopharm Chemical Reagent, catalog number: 10009228 ) (ice cold)
- 70% ethanol (ice cold)
- 7.5 M ammonium acetate (Sigma-Aldrich, catalog number: A1542-250G )
- Chloroform: isoamyl alcohol (24:1) (Sinopharm Chemical Reagent, catalog number: chloroform, 10006818 ; isoamyl alcohol, 10003218 )
- ddH2O (sterile)
- 5x Loading buffer with GelRed (Shanghai Generay Biotech, catalog number: GR0205-500 )
- Cetyltrimethylammonium bromide/Hexadecyl trimethyl-ammonium bromide (CTAB) (Thermo Fisher Scientific, catalog number: ICN19400480 )
- Ethylenediaminetetraacetic acid (0.5M solution/pH 8.0) (EDTA) (Thermo Fisher Scientific, catalog number: BP2482-500 )
- Polyvinyl pyrrolidone, MW 40,000 (PVP 40)
- NaCl (Sigma-Aldrich, catalog number: S5886-500G )
- HCl (Sigma-Aldrich, catalog number: 258148 )
- Tris base (Thermo Fisher Scientific, catalog number: BP152-1 )
- Boric acid (Sigma-Aldrich, catalog number: B0394-100G )
- Agarose (Biowest, catalog number: 111860 )
- Hi-DiTM formamide (Thermo Fisher Scientific, Applied BiosystemsTM, catalog number: 4311320 )
- GeneScan 500Liz (Thermo Fisher Scientific, Applied BiosystemsTM, catalog number: 4322682 )
- CTAB buffer (see Recipes)
- 1 M Tris (pH 8.0) (see Recipes)
- TE buffer (see Recipes)
- 5x TBE (Tris-Borate-EDTA) buffer (Stock) (see Recipes)
- 1% agarose gel (see Recipes)
- Taq PCR mastermix (Tiangen, catalog number: KT201 )
Equipment
- Mortar and pestle
- Microcentrifuge (Eppendorf, model: 5427R )
- Water bath (IKA, model: HB10 )
- PCR thermal cycler (EASTWIN, model: ETC-811 )
- Agarose gel electrophoresis system [include PowerPacTM Basic Power Supply for electrophoresis (Bio-Rad Laboratories, catalog number: 1645050 )]
- Genetic Analyzer (Invitrogen, model: ABI PRISM 3100 )
Software
- GeneMarker version 2.4.0 (http://www.softgenetics.com/GeneMarker.html)
- FSTAT version 2.9.3 (http://www2.unil.ch/popgen/softwares/fstat.htm)
- GenAlEx version 6.2 (http://biology-assets.anu.edu.au/GenAlEx/Download.html)
- STRUCTURE version 2.2 (http://pritchardlab.stanford.edu/software/structure2_2.html)
- NewHybrids version 1.1 beta (http://ib.berkeley.edu/labs/slatkin/eriq/software/software.htm#NewHybs)
Procedure
- DNA extraction and polymerase chain reaction
Total DNA was extracted using a modified cetyltrimethylammonium bromide (CTAB) method (Doyle, 1991). Ten pairs of SSR markers were developed by Duan and Zhang (2014) (AC30, CT12, CT59, CT113, CT17, CT60, CT48, CT135, CT142, CT91) from M. pubescens were selected for testing the gene flow between M. pubescens var. alba and M. kwangtungensis. The forward of each primer pair was labeled with fluorochrome (FAM, HEX, ROX, TAMRA). The information of the primers is shown in Table 1.
Table 1. Characteristics of the 10 pairs of SSR primers employed in this study. Presented for each locus are the forward (F) and reverse (R) primer sequences, repeat motif, size of the original fragment (bp), annealing temperature (Ta), and GenBank accession number. Td, touchdown.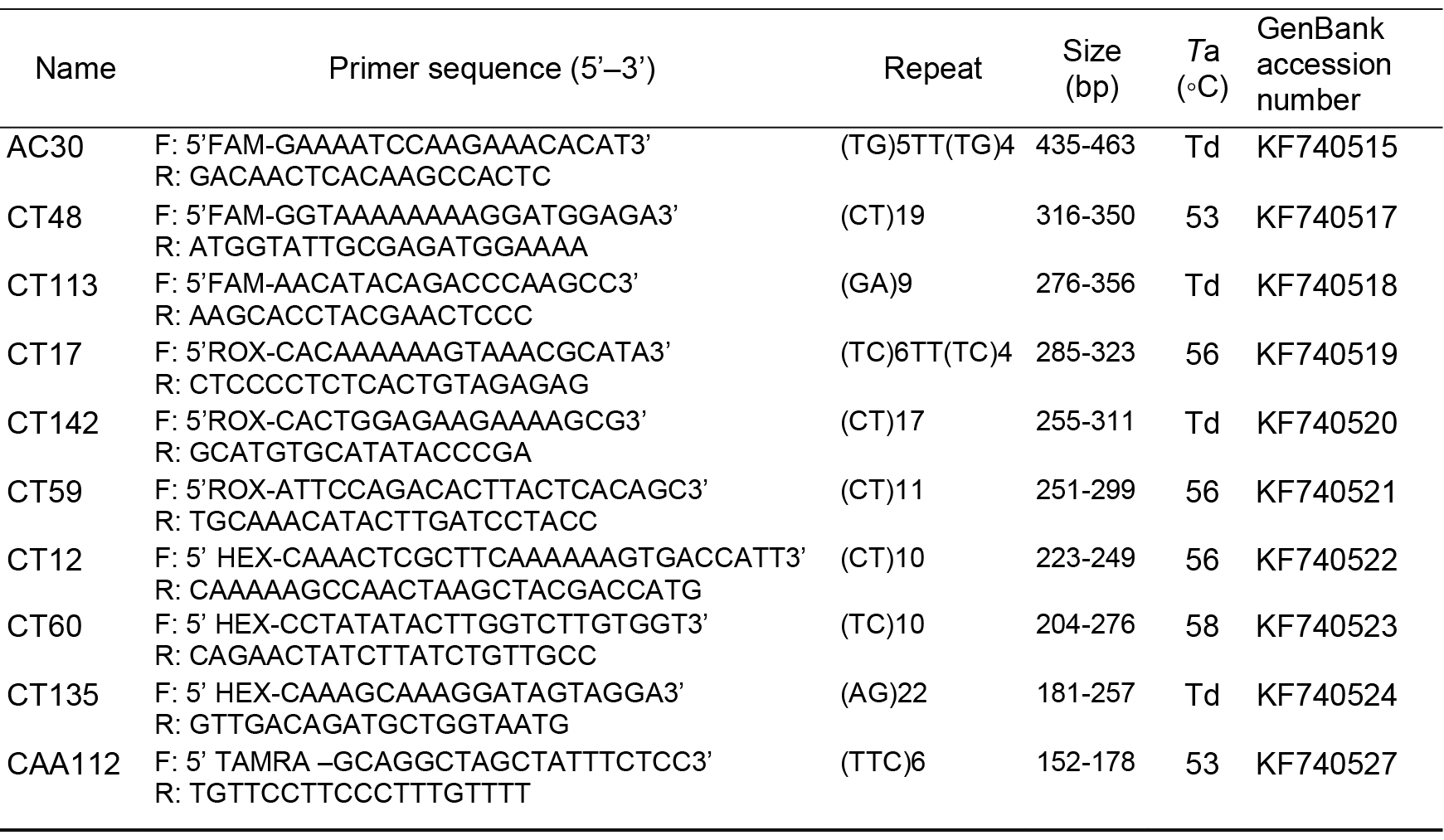
DNA amplifications were carried out in 10 μl reaction mixtures:
5 μl Taq PCR mastermix
0.2 μM of each primer
3.6 μl ddH2O
20-40 ng of genomic DNA
The amplification conditions were as follows:- 94 °C, 4 min (initial denaturation)
- 94 °C, 30 sec (denaturation)
- 60 °C, 30 sec (annealing)
- 72 °C, 30 sec (extension)
7 cycles from step (b) to (d) - 94 °C, 30 sec (denaturation)
- 53 °C, 30 sec (annealing)
- 72 °C, 30 sec (extension)
28 cycles from step (e) to (g) - 72 °C, 10 min (final extension)
- Keep sample at 4 °C until further processing
The PCR products were determined on an ABI PRISM 3100 Genetic Analyzer in the Sangon Biotech (Shanghai) Co., Ltd., and 1 μl products (10-fold diluted) were mixed with 15 μl Hi-DiTM Formamide probe, which included 2% GeneScan 500Liz. The amplified SSR fragment data were collected using GeneMarker version 2.4.0 software. - Data analysis
The software FSTAT 2.9.3 (Goudet, 2001) and GenAlEx 6.2 (Peakall and Smouse, 2006) were used for calculating the alleles and private alleles, respectively. We used the program STRUCTURE 2.2 (Pritchard et al., 2000; Falush et al., 2003 and 2007) for population genetic structure analysis and hybrid detection. This method implements a Bayesian model-based clustering approach. We supposed all the loci were independent, following the Hardy–Weinberg equilibrium and linkage equilibrium. The program was run 10 times for each K-value, ranging 1-5, with 100,000 replicates for burn-in and 40,000 iterations. Out of the five runs for k = 2, the run with the highest likelihood value was selected to assign the posterior membership coefficients (Evanno et al., 2005).
The entire data set is arranged as a matrix in a single file, in which the data for individuals are in rows, and the loci are in columns. For a diploid organism, data for each individual can be stored either as 2 consecutive rows, where each locus is in one column, or in one row, where each locus is in two consecutive columns. Missing data should be indicated by a number that doesn’t occur elsewhere in the data.
The software NewHybrids version 1.1 beta (Anderson and Thompson, 2002) was used to identify hybrids in the individuals sampled. The program calculates the posterior probability that fall into each of the six genotype categories, Parent 1, Parent 2, F1 hybrids, F2 hybrids, back-cross generation to Parent 1, and back-cross generation to Parent 2.
For FSTAT, GenAlex and NewHybrids, we used the default value in each software. For STRUCTURE, we chose admixture model, and set the parameters as we mentioned in the original article (Luo et al., 2015), other parameters we used the default values.
Representative data
A representative example of data can be found in our published work (Luo et al., 2015). Figure 1 shows genotype class analysis of all sampled individuals based on the programs NewHybrids and STRUCTURE using SSR data.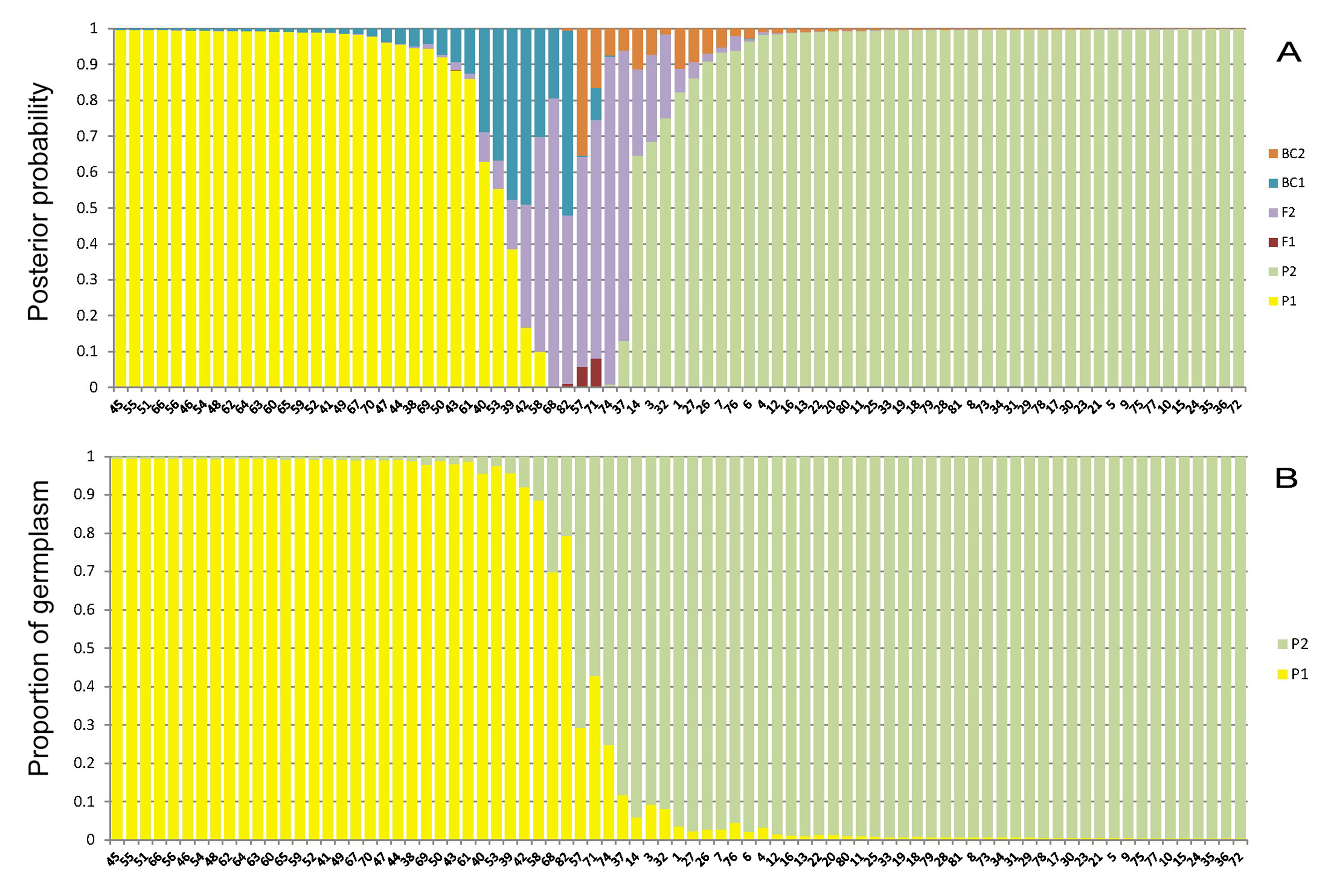
Figure 1. Genotype class analysis of all sampled individuals based on the programs NewHybrids (A) and STRUCTURE (B) using SSR data. A. NewHybrids calculated posterior probability that falls into each of six genotype categories: P1 (Parent 1), P2 (Parent 2), F1 (F1 hybrids), F2 (F2 hybrids), BC 1 (backcross generation to Parent 1), BC 2 (backcross generation to Parent 2). B. Structure of 81 accessions based on 13 SSR loci (K = 2). Each bar represents one individual, P1 = M. kwangtungensis, P2 = M. pubescens var. alba.
Recipes
- CTAB buffer, 100 ml
2.0 g CTAB
10.0 ml 1 M Tris (pH 8.0)
4.0 ml 0.5 M EDTA (pH 8.0)
28.0 ml 5 M NaCl
40.0 ml H2O
1 g PVP 40
Adjust all to pH 8.0 and make up to 100 ml with ddH2O. - 1 M Tris pH 8.0, 1 L
121.1 g Tris
700 ml ddH2O
Dissolve Tris in ddH2O and bring to 900 ml.
Adjust pH to 8.0 by adding 40-50 ml of concentrated HCl. - TE buffer for 100 ml
10 mM 1 ml of 1 M Tris (pH 8.0)
1 mM 0.2 ml of 0.5 M EDTA - 5x TBE (Tris-Borate-EDTA) buffer (Stock)
54 g Tris base
27.5 g boric acid
20 ml 0.5 M EDTA (pH 8.0)
Adjust pH to 8.3 by HCl.
This stock solution can be diluted to 1x prior to use in electrophoresis. - 1% agarose gel
Dissolve 1 g agarose in 100 ml 1x TBE
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31000109, 31370269, 31170184, U1202261) and the International Foundation for Science (AD/22476). This protocol was modified from our previous work, Luo, Z. L., Duan, T. T., Yuan, S., Chen, S., Bai, X. F., and Zhang, D. X. (2015) Reproductive isolation between sympatric sister species, Mussaenda kwangtungensis and M. pubescens var. alba. J Integr Plant Biol 57: 859-870.
References
- Anderson, E. C. and Thompson, E. A. (2002). A model-based method for identifying species hybrids using multilocus genetic data. Genetics 160(3): 1217-1229.
- Doyle, J. (1991). DNA protocols for plants-CTAB total DNA isolation. In: Hewitt, G. M. and Johnston, A. (eds). Molecular techniques in taxonomy. Springer, 283-293.
- Duan, T. and Zhang, D. (2014). Fourteen additional microsatellite markers for Mussaenda pubescens and cross-species amplification. J Genet 93(2): e44-47.
- Evanno, G., Regnaut, S. and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol Ecol 14: 2611-2620.
- Falush, D., Stephens, M. and Pritchard, J. K. (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164(4): 1567-1587.
- Falush, D., Stephens, M., and Pritchard, J. K. (2007). Inference of population structure using multilocus genotype data: Dominant markers and null allele. Mol Ecol Notes 7: 574-578.
- Goudet, J. (2001). FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3) (updated from Goudet, J. (1995). FSTAT (version 1.2): a computer program to calculate F-statistics. J. Hered. 86: 485-486). Available at: http://www.unil.ch/izea/softwares/fstat.html.
- Luo, Z. L., Duan, T. T., Yuan, S., Chen, S., Bai, X. F., and Zhang, D. X. (2015) Reproductive isolation between sympatric sister species, Mussaenda kwangtungensis and M. pubescens var. alba. J Integr Plant Biol 57: 859-870.
- Peakall, R. and Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research--an update. Bioinformatics 28(19): 2537-2539.
- Pritchard, J., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945-959
- Shaw, J., Lickey, E. B., Schilling, E. E. and Small, R. L. (2007). Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am J Bot 94(3): 275-288.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Luo, Z., Duan, T., Yuan, S., Chen, S., Bai, X. and Zhang, D. (2016). Identification of Natural Hybrids by SSR Markers in Mussaenda. Bio-protocol 6(13): e1853. DOI: 10.21769/BioProtoc.1853.
Category
Plant Science > Plant molecular biology > DNA > Genotyping
Molecular Biology > DNA > Genotyping
Molecular Biology > DNA > PCR
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











