- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Extraction and Profiling of Plant Polar Glycerol Lipids
Published: Vol 6, Iss 12, Jun 20, 2016 DOI: 10.21769/BioProtoc.1849 Views: 12102
Reviewed by: Marisa RosaAgnieszka ZienkiewiczAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Sorghum bicolor Extracellular Vesicle Isolation, Labeling, and Correlative Light and Electron Microscopy
Deji Adekanye [...] Jeffrey L. Caplan
Oct 5, 2024 2117 Views

A New Approach to Detect and Semi-quantify All Molecular Species and Classes of Anionic Phospholipids Simultaneously in Plant Samples
Manon Genva [...] Laetitia Fouillen
Apr 20, 2025 1730 Views

PhosphoLIMBO: An Easy and Efficient Protocol to Separate and Analyze Phospholipids by HPTLC From Plant Material
Louise Fougère [...] Yohann Boutté
Sep 5, 2025 1343 Views
Abstract
This protocol describes a method to extract total polar glycerol lipids from plant materials, followed by mass spectrometry profiling. Different glycerol lipid classes can be distinguished by their head-groups, which can be profiled automatically and quantitatively by a triple quadrupole mass spectrometry in multiple reaction monitoring (MRM) mode with an autosampler. Comparing with other established methods, such as thin layer chromatography (TLC) separation followed by Gas spectrometry (GC) analysis, this method requires little effort in sample preparation and separation, while the resolution is not limited to general lipid classes but at side chain level. This method was described and used successfully to profile plant lipids changes under freezing stress in Welti et al. (2002).
Keywords: Mass spectrometryMaterials and Reagents
- PYREXTM screw cap culture tubes with PTFE lined phenolic caps (Thermo Fisher Scientific, catalog number: 14-933D )
- Disposable glass pipettes (Thermo Fisher Scientific, Fisher ScientificTM, catalog number: 13-678-20C )
- Autosampler vials (Sigma-Aldrich, catalog number: 29133-U ) with pre-cut cap (Sigma-Aldrich, catalog number: 29494-U )
- TLC Silica gel 60 plate (Merck)
- 1 ml syringe
- Injection loop
- 4-6 weeks old Arabidopsis rosette leaves
Note: Other plant materials can be used, additional precautions may be needed (see Notes). - Isopropanol (Thermo Fisher Scientific, catalog number: A416 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9333 )
- Chloroform (Thermo Fisher Scientific, catalog number: C607 )
- Methanol (Thermo Fisher Scientific, Fisher ScientificTM, catalog number: A456 )
- Acetic acid
- Iodine
- Primuline
- Acetone
- Nitrogen gas
- Butylated hydroxytoluene (BHT) (Sigma-Aldrich, catalog number: B1378 )
- 7.5 M ammonium acetate solution (NH4Ac) (Sigma-Aldrich, catalog number: A2706 )
- Phospholipid (PL) standard [kindly provided by Dr. Ruth Welti, (Kansas State University)]
Note: An example PL standard list is shown in Table 1. - Monogalactosyldiacylglycerol (MGDG), hydrogenated (Matreya, catalog number: 1058 )
- Digalactosyldiacylglycerol (DGDG), hydrogenated (Matreya, catalog number: 1059 )
Note: The actual composition of MGDG and DGDG standards need to be determined experimentally, such as transmethylation followed by GC analysis (Politz et al., 2013). - Isopropanol solution with BHT (see Recipes)
- Lipid extraction solution (see Recipes)
- KCl solution (see Recipes)
- Solution B (see Recipes)
- Galactolipid (GL) standard (see Recipes)
- Phospholipid (PL) standard (see Recipes)
Equipment
- Water bath (Thermo Fisher Scientific, model: Isotemp2340 )
- Water bath shaker (Labnet International, model: SWB5050 )
- Nitrogen evaporator N-EVAP (Organomation, model: 111 )
- Fume hood
- Ultraviolet light (360 nm)
- Oven (Thermo Fisher Scientific, Fisher ScientificTM, model: 13-247-650G )
- API 4000 LC-MS/MS system (AB SCIEX) with TurboIonspray probe and HTC PAL autosampler (LEAP Technologies)
Table 1. Example of PL standard list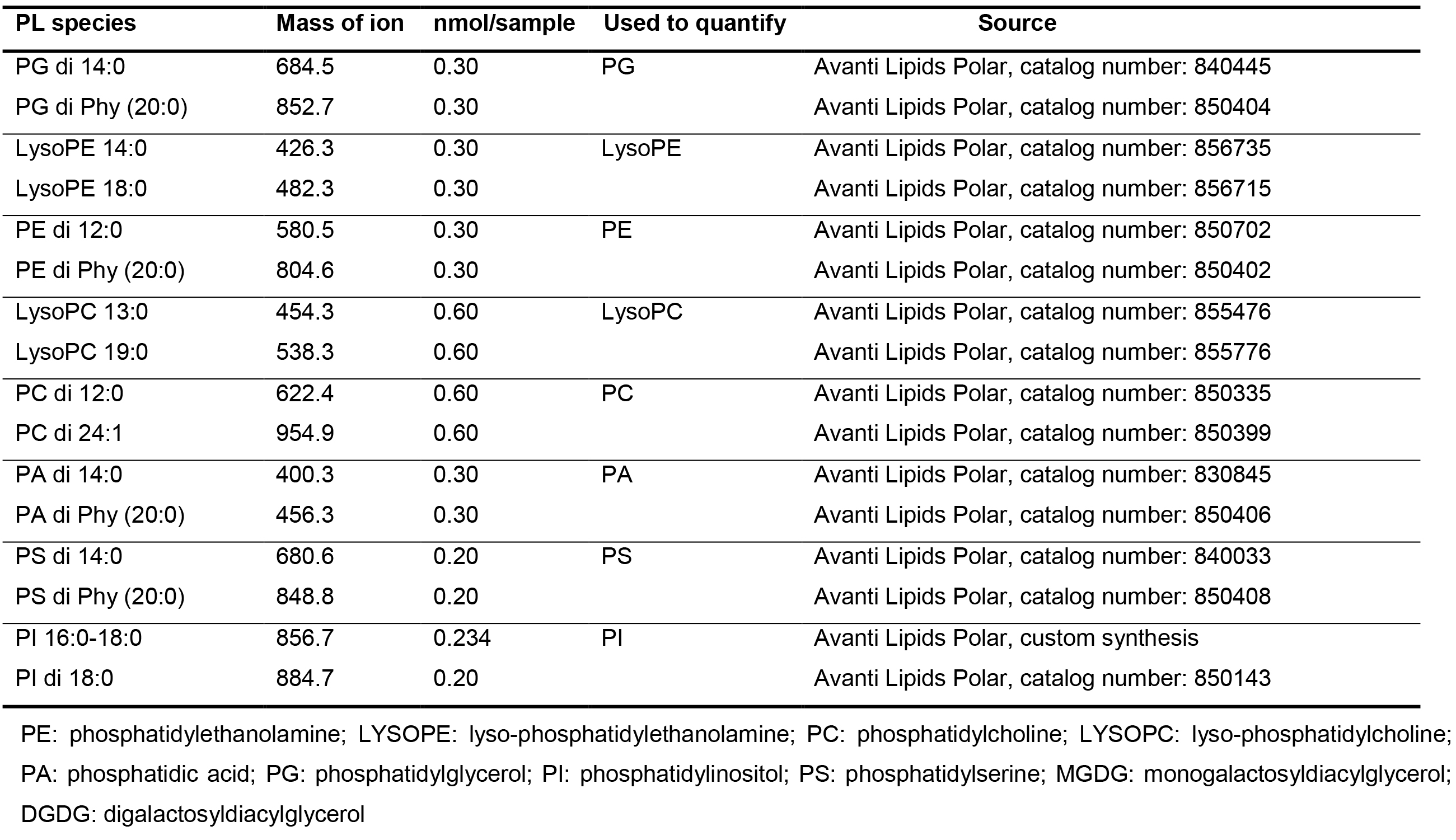
Software
- Analyst 1.5.1 (AB SCIEX)
Procedure
- Total lipid extraction (Figure 1)
- Aliquot 3 ml preheated isopropanol (75 °C with 0.01% BHT) into glass tubes (50 ml, 25 x 150 mm tubes with Teflon-lined screw caps are recommended).
- Fresh Arabidopsis leaves were harvested and immediately soaked in hot isopropanol for at least 15 min in 75 °C water bath to inactivate phospholipase D. To achieve higher reliability and repeatability, 5-30 mg delipidated “dry weight” as measured in step A10 is recommended (~2-8 fully developed leaves to start with). Samples were cooled to room temperature after incubation. Isopropanol treated samples are stable at room temperature within a day.
- Add 1.5 ml chloroform and 0.6 ml water to cooled samples, mix by vortexing. Incubate samples in water bath shaker at room temperature, 50 rpm for 1 h.
- Transfer extracted lipids (disregard potential phase separation) with a glass Pasteur pipettes to another glass collection tube.
- Add 4 ml lipid extraction solution to leaf samples and incubate in 25 °C water bath shaker for 30 min. Transfer extracts to the same collection tube.
- Repeat step A5 to extract samples with lipid extraction solution until all leaf samples become white. It is recommended to extract each sample the same number of times. 5 extractions (including isopropanol in step A2) are usually needed. Old leaves are harder to be extracted completely, longer incubation time in steps A2, A3 and A5 and more extraction steps may help.
- Add 1 ml KCl solution to the combined extract (~20 ml), mix thoroughly by vortex, centrifuge at ~200 x g for 10 min, discard upper phase carefully.
- Add 2 ml water, mix by vortex, centrifuge at ~200 x g for 10 min, discard upper phase carefully.
- Dry lipid samples completely in the nitrogen evaporator, mild heating (<50 °C) may be used to accelerate the process. Dried lipids can be stored at -80 °C with cap tightly closed. An aliquot of lipids can be resolved by thin layer chromatography (TLC) for quality check (Figure 2). Briefly, total lipids corresponding to ~0.2-1 mg DW were spotted on the TLC Silica gel 60 plate and resolved by chloroform:methanol:acetic acid:H2O = 85:15:12.5:3.5. The plate was left dry completely in the fume hood before staining by iodine vapor and examining by eye or 0.01% primuline in acetone/H2O (60:40; v/v) and examining the plates under ultraviolet (360 nm) light.
- Dry extracted leaves in oven at 105 °C overnight. Weigh to get “dry weight”.
- Aliquot 3 ml preheated isopropanol (75 °C with 0.01% BHT) into glass tubes (50 ml, 25 x 150 mm tubes with Teflon-lined screw caps are recommended).
- Lipid profiling by ESI-MS/MS and data processing
- Reconstitute dried lipid samples (step A9) in chloroform, cap tightly before use to avoid evaporation. For easy sample preparation, lipid samples were diluted to the same concentration according to dry weight. We usually use 250 µl chloroform per mg dry weight.
- Each sample contains 840 µl solution B, 5 µl PL standard, 3 µl GL standard and lipid sample corresponding to 0.1-0.2 mg dry weight (25-50 µl), add chloroform to 1,200 µl in a 2 ml snap seal autosampler vial, cap with pre-cut vial caps.
- For every 10 lipid samples, only one “internal standard (IS)” vial is used for machine correction/contamination control. Chloroform is used in place of lipid samples in this vial.
- Load the autosampler in the following format: IS1, sample1-10, IS2, sample11-20 and so on. “Blank” samples are recommended between different genotypes, which contains only solution B and chloroform.
- Profile lipid samples by ESI-MS/MS. Detailed information can be found in Welti et al. (2002). The parameters we used on API 4000 are listed in section C. Optimization is needed on different setups, such that fewer or more parameters can be adjusted on older or newer machines. A representative phosphatidylethanolamine (PE) profiling result is attached (Figure 3).
- Data were extracted by Analyst 1.5.1 with necessary corrections, including background subtraction and smooth. For each lipid class, multiple (usually 2) novel lipid species were used as internal standards (Table 1), linear interpolation according to molecular weight of each lipid species was used for internal standard peak area normalization. The amount of each lipid species was calculated according to sample peak area compared with normalized internal standard peak area. Five or more biological repeats are recommended, Student’s t-test is used for statistical analysis.
- Lipid data may be presented as absolute amount (nmol/mg “dry weight”, recommended) or percentage of total lipids (PL and GL).
- Reconstitute dried lipid samples (step A9) in chloroform, cap tightly before use to avoid evaporation. For easy sample preparation, lipid samples were diluted to the same concentration according to dry weight. We usually use 250 µl chloroform per mg dry weight.
- Lipid profiling parameters on API 4000
API 4000 LC-MS/MS system equipped with TurboIonSpray probe and HTC PAL autosampler was used for lipid profiling. The source temperature was 100 °C, interface heater was on, IonSpray Voltage was set to 5.5 kV, nitrogen gas was used for both source and curtain gas. The scan resolution was set to 0.1Da. The samples were introduced using the autosampler with 1 ml syringe and injection loop for full lipid profiling. The samples were presented to the TurboIonSpray probe by direct infusion at 0.03 ml/min. The scanning parameters for each lipid class were listed in Table 2.
Table 2. Scanning parameters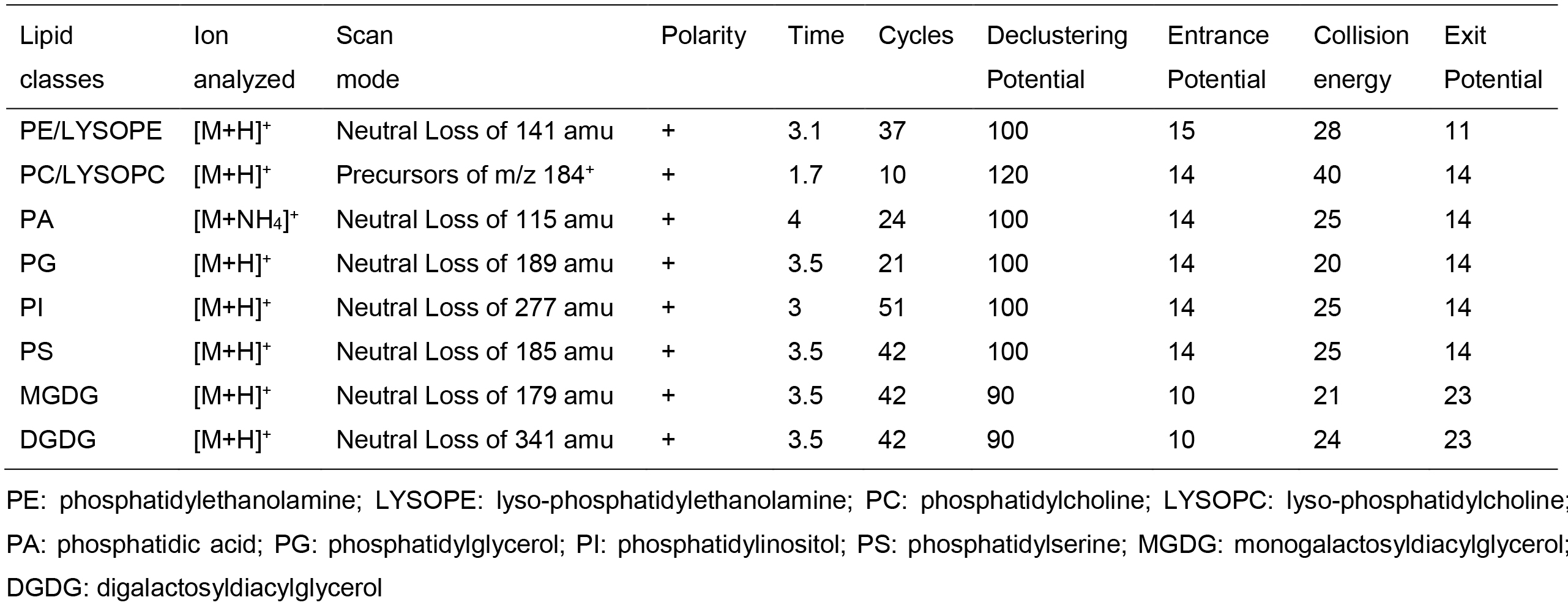
Representative data
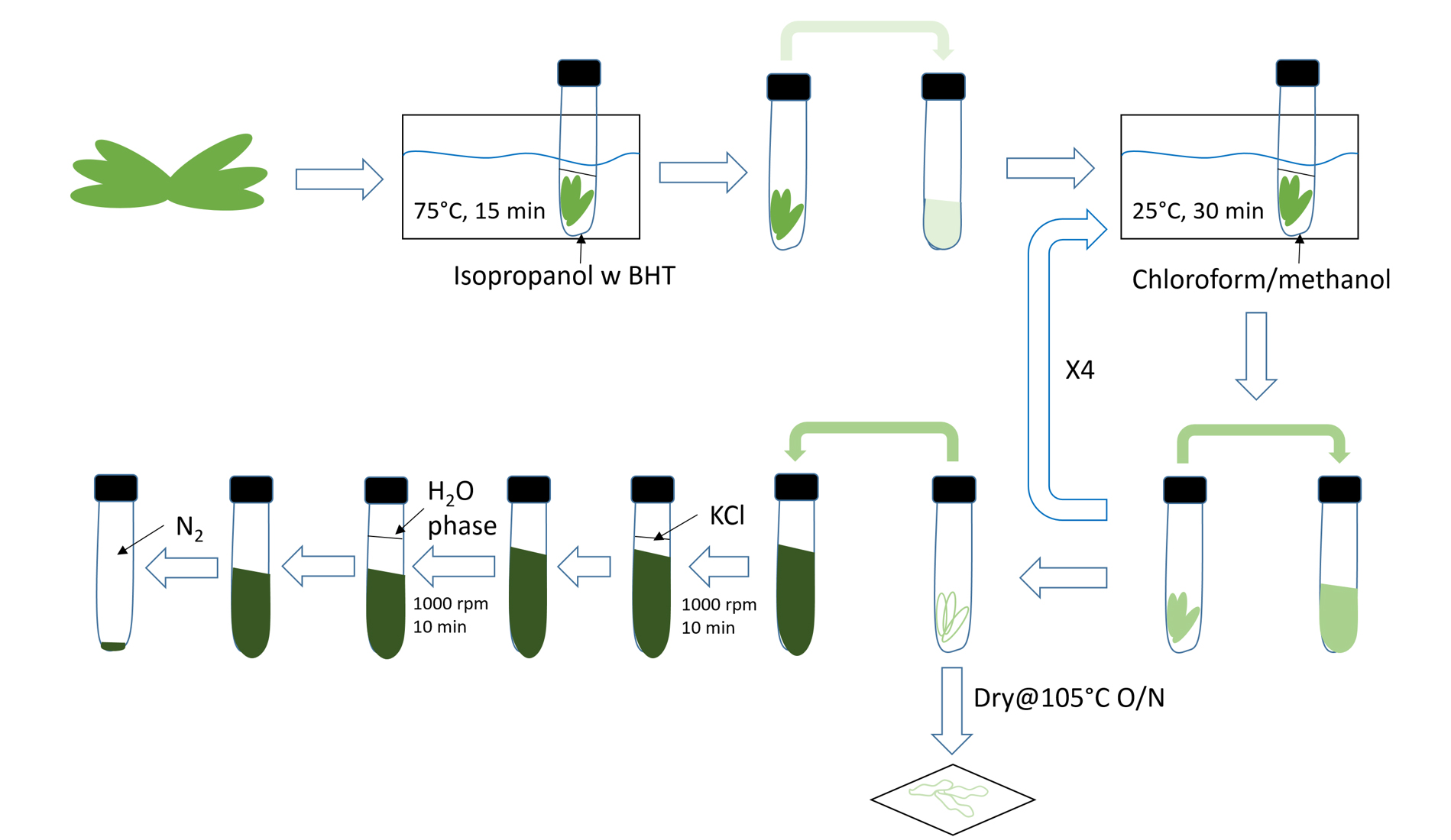
Figure 1. Illustration of lipid extraction workflow. 1,000 rpm (~200 x g).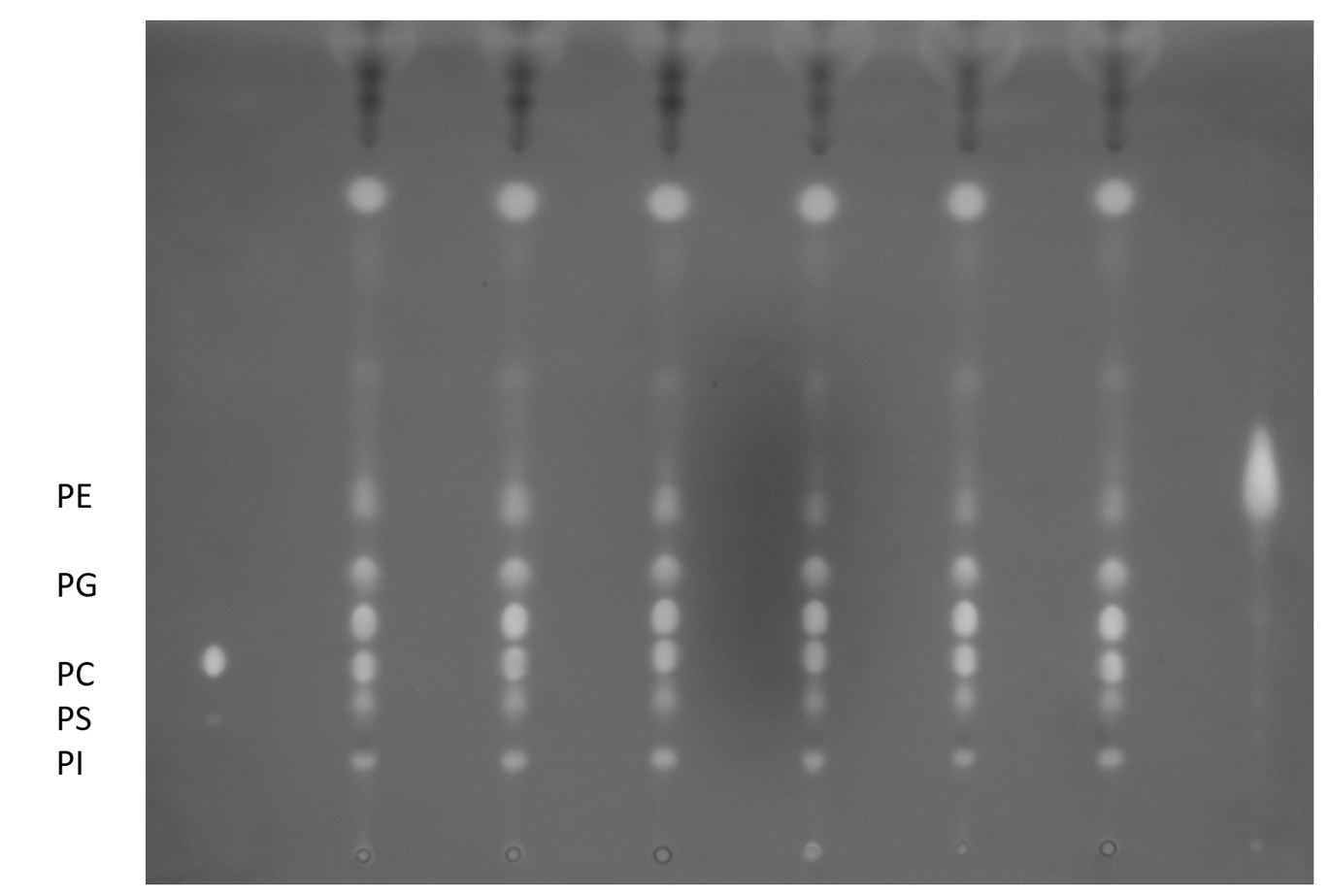
Figure 2. TLC separation of phospholipids. Lipids extracted in section A can be resolved on a TLC plate. The mobile phase used here is chloroform:methanol:acetic acid:H2O = 85:15:12.5:3.5 (v/v).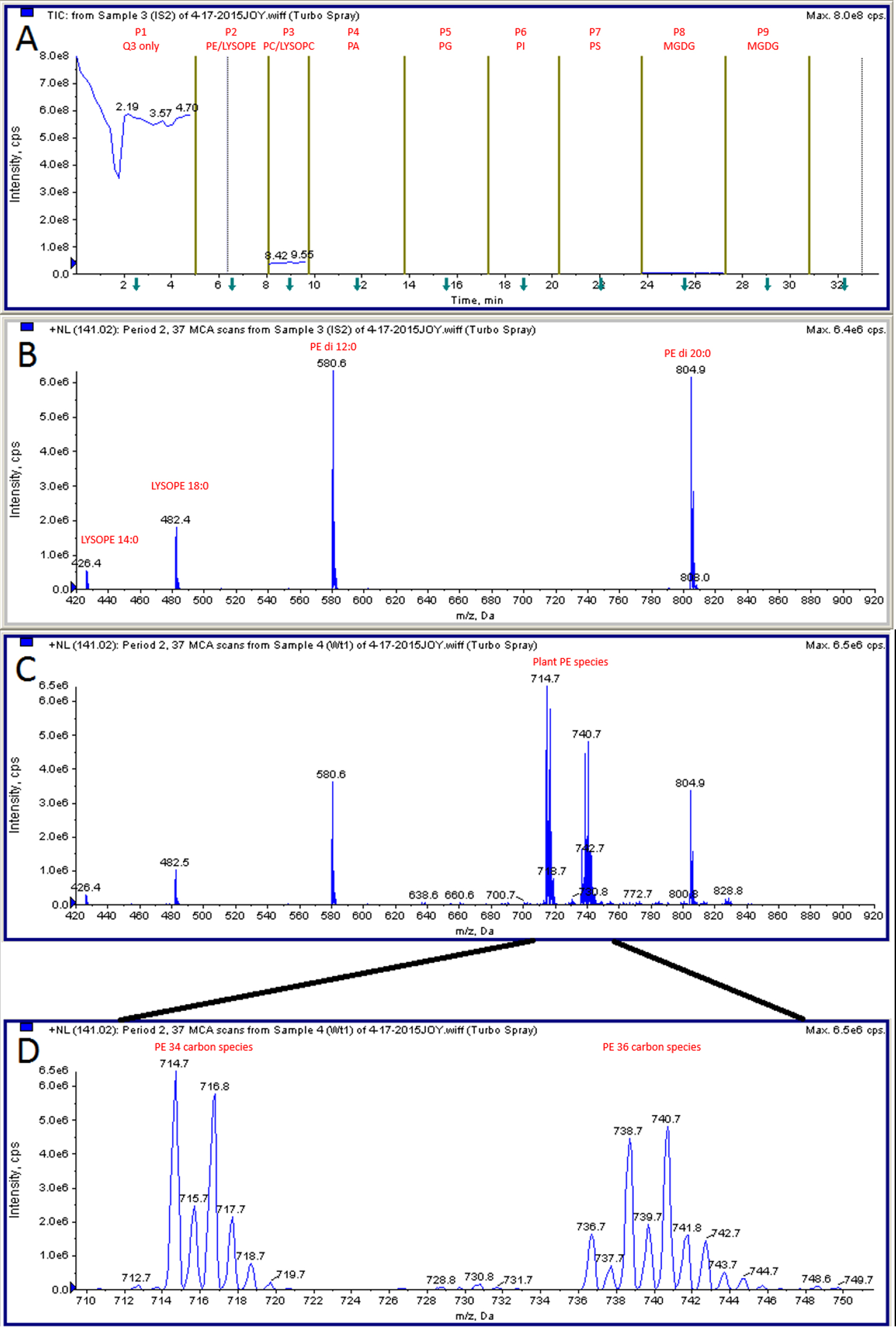
Figure 3. Representative MS data. A. Different lipid classes were profiled in tandem periods. Period 1 is for control purpose and not used in calculation. B. PE profile in IS sample. C. PE profile in plant sample. D. Enlarged view of plant PE species.
Notes
- This Method can be easily adapted to roots, flowers, stems and whole siliques.
- For Arabidopsis seed samples, the reproducibility of “dry weight” is highly dependent on seed stage and loss of samples during transferring lipid extracts. In this case, the percentage normalization is recommended.
- For materials that are hard to extract completely, tissue disruption in liquid nitrogen before extraction is recommended. In this case, a centrifugation step (e.g., ~200 x g, 10 min) is needed each time before transferring lipid extracts.
Recipes
- Isopropanol solution with BHT
0.01% (w/v) BHT (~0.5 mM)
Preheat at 75 °C
Note: stable at room temperature - Lipid extraction solution
Chloroform:methanol = 2:1 (v/v), with 0.01% BHT
Note: stable if capped tightly at room temperature - KCl solution
1 M KCl
Note: stable at room temperature - Solution B
Methanol:NH4Ac (300 mM) = 20:1(v/v)
Note: stable at room temperature - Galactolipid (GL) standard
Note: store at -80 °C, thaw and mix thoroughly before use
Diluted in chloroform:methanol:H2O = 35:65:8 [(v/v), or close to]
MGDG (~0.5 nmol/µl)
DGDG (~0.25 nmol/µl) - Phospholipid (PL) standard
Note: store at -80 °C, thaw and mix thoroughly before use
Diluted in chloroform:methanol:H2O = 35:65:8 [(v/v), or close to]
See Table 1 for an example of PL standard content
Acknowledgments
This protocol was adapted from Welti et al. (2002). Dr. Welti also provided help in lipid standards, data processing and analysis. Work was supported by the U.S. Department of Energy (DOE) Grant DE-AR0000202 and by the National Science Foundation Grants MCB-1412901 and DBI-1427621.
References
- Politz, M., Lennen, R. and Pfleger, B. (2013). Quantification of bacterial fatty acids by extraction and methylation. Bio-protocol 3(21): e950.
- Welti, R., Li, W., Li, M., Sang, Y., Biesiada, H., Zhou, H. E., Rajashekar, C. B., Williams, T. D. and Wang, X. (2002). Profiling membrane lipids in plant stress responses. Role of phospholipase D alpha in freezing-induced lipid changes in Arabidopsis. J Biol Chem 277(35): 31994-32002.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Liu, Y. and Wang, X. (2016). Extraction and Profiling of Plant Polar Glycerol Lipids. Bio-protocol 6(12): e1849. DOI: 10.21769/BioProtoc.1849.
Category
Biochemistry > Lipid > Lipid isolation
Biochemistry > Lipid > Lipid measurement
Plant Science > Plant biochemistry > Lipid
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link