- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Whole-mount Enteroid Proliferation Staining
Published: Vol 6, Iss 12, Jun 20, 2016 DOI: 10.21769/BioProtoc.1837 Views: 12131
Reviewed by: Ivan ZanoniAchille BroggiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation and Quantification of Mouse γδT-cells in vitro and in vivo
Isha Rana [...] Colin Jamora
Sep 5, 2021 5007 Views

Sample Preparation and Integrative Data Analysis of a Droplet-based Single-Cell ATAC-sequencing Using Murine Thymic Epithelial Cells
Tatsuya Ishikawa [...] Taishin Akiyama
Jan 5, 2023 2415 Views

Proliferation Assay Using Cryopreserved Porcine Peripheral Mononuclear Cells Stimulated With Concanavalin A and Analyzed With FCS ExpressTM 7.18 Software
Marlene Bravo-Parra [...] Luis G. Giménez-Lirola
Jun 5, 2025 2620 Views
Abstract
Small intestinal organoids, otherwise known as enteroids, have become an increasingly utilized model for intestinal biology in vitro as they recapitulate the various epithelial cells within the intestinal crypt (Mahe et al., 2013; Sato et al., 2009). Assessment of growth dynamics within these cultures is an important step to understanding how alterations in gene expression, treatment with protective and toxic agents, and genetic mutations alter properties essential for crypt growth and survival as well as the stem cell properties of the individual cells within the crypt. This protocol describes a method of visualization of proliferating cells within the crypt in three dimensions (Barrett et al., 2015). Whole-mount proliferation staining of enteroids using EdU incorporation enables the researcher to view all proliferating cells within the enteroid as opposed to obtaining growth information in thin slices as would be seen with embedding and sectioning, ensuring a true representation of proliferation from the stem cell compartment to the terminally differentiated cells of the crypt.
Materials and Reagents
- 12-well MatTek plate with 1.5 coverslip thickness (MATTEK, catalog number: P12G-1.5-14-F )
- 18 G non-flexible stainless steel oral gavage needle (Cadence, Inc., catalog number: 7906 )
- 10 ml syringe (Thermo Fisher Scientific, Fisher ScientificTM, catalog number: 14-823-2A )
- 50 ml sterile Falcon tubes (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 339653 )
- Sterile 12-well cell culture dish (Sigma-Aldrich, catalog number: CLS3513 )
- 15 ml sterile Falcon tubes (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 339651 )
- Pre-chilled sterile pipette tips
Notes:- Any tips that are sterile and can be used with the lab’s pipettors.
- These tips can be placed in a -20 °C freezer for 30 min or in a 4 °C refrigerator one hour prior to plating.
- 70 μM cell strainer (Thermo Fisher Scientific, Fisher ScientificTM, catalog number: 08-771-2 )
- C57BL/6 mouse (male or female, 6-8 weeks of age)
- Isofluorane (Allivet, catalog number: 50562 )
- Ice cold sterile 1x DPBS without calcium or magnesium (Thermo Fisher Scientific, GibcoTM, catalog number: 14190-235 )
- UltraPure EDTA (Thermo Fisher Scientific, InvitrogenTM, catalog number: 15576-028 )
- Sucrose (Sigma-Aldrich, catalog number: S0389 )
- D-Sorbitol (Sigma-Aldrich, catalog number: S1876 )
- Matrigel® basement membrane matrix (Corning, catalog number: 356237 )
Note: Thaw at 4 °C the night before use. - Epidermal growth factor (EGF) (R&D Systems, catalog number: 2028-EG-200 )
- Noggin (R&D Systems, catalog number: 1967-NG-025/CF )
- R-spondin (R&D Systems, catalog number: 3474-RS-050 )
- Wnt3A (R&D Systems, catalog number: 1324-WN-010 )
- Advanced DMEM/F12 (Thermo Fisher Scientific, GibcoTM, catalog number: 12634-010 )
- L-glutamine (Thermo Fisher Scientific, GibcoTM, catalog number: 25030-081 )
- Penicillin-streptomycin (Thermo Fisher Scientific, GibcoTM, catalog number: 15140-148 )
- 1 M HEPES (pH 7.0-7.6, Sterile filtered) (Sigma-Aldrich, catalog number: H0887 )
- N2 supplement (R&D Systems, catalog number: AR003 )
- B27 supplement (Thermo Fisher Scientific, GibcoTM, catalog number: 17504-044 )
- Fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10437-028 )
- Click-iT EdU cell proliferation assay (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: C10337 )
- Paraformaldehyde (Sigma-Aldrich, catalog number: P6148 )
- Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A8531 )
- Triton X-100 (Sigma-Aldrich, catalog number: X100 )
- TO-PRO-3 Iodide (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: T3605 )
- Chelation buffer, prepare fresh (see Recipes)
- Shaking buffer (see Recipes)
- Minigut culture media (see Recipes)
- 2% Formaldehyde (see Recipes)
Equipment
- Scissors for dissection (Fisher Scientific, catalog number: 08-951-20 )
- Sorvall Legend X1R Centrifuge (or other large refrigerated centrifuge)
- 37 °C, 5% CO2 cell culture incubator (Thermo Fisher Scientific, model: HERAcell 150i )
- LSM 510 META Inverted (or other inverted laser scanning) confocal microscope with 647 (TO-PRO-3) and 488 (EdU) emission filters
Software
- ImageJ Image Analysis Software
Procedure
- Harvest and culturing of small intestinal enteroids
- Anesthetize and sacrifice mouse with isofluorane followed by cervical dislocation.
- Dissect out approximately 10 cm of the proximal intestine (duodenum, near stomach, Figure 1A).
- Fill a 10 ml syringe with ice cold PBS and attach the oral gavage needle to the syringe.
- Insert the gavage needle into the proximal intestine (from step A2). Flush the intestine with ice cold PBS (Figure 1B).
- Cut the intestine open lengthwise (Figure 1C) and cut into 1 cm pieces and transfer into 5 ml ice cold PBS in a 15 ml conical tube (Figure 1D). Vortex for 3 sec to remove loose villi.
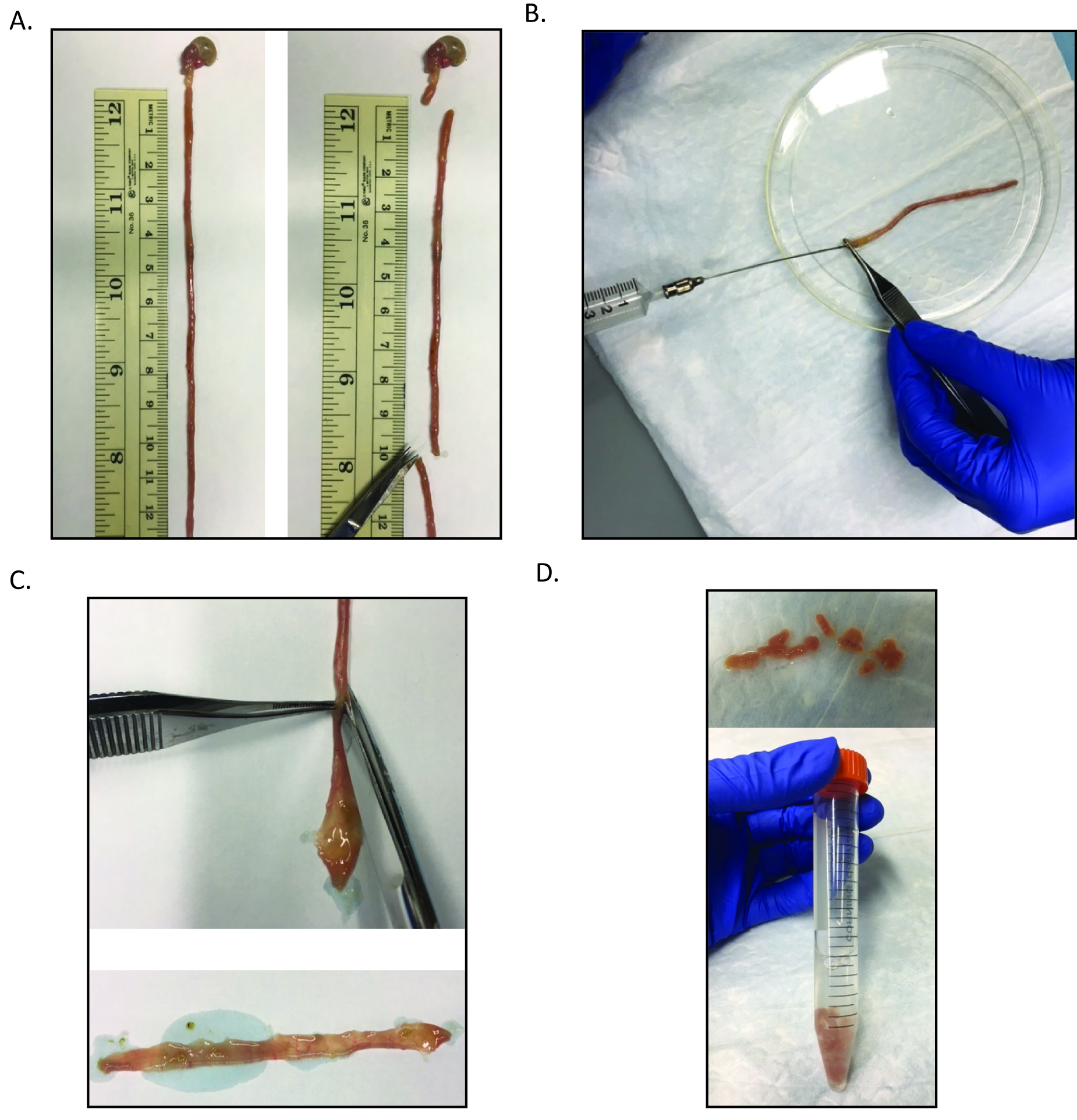
Figure 1. Processing of the small intestine for crypt isolation. A. After removal of the small intestine, the most proximal 10 cm should be collected for further processing. B. Then this section should be flushed with ice cold PBS using the gavage needle attached to the 10 ml syringe. C. The intestinal section is then bisected. D. The bisected intestine is then cut into approximately 1 cm sections and is transferred to a 15 ml conical tube containing 5 ml ice cold PBS. - Pipette off PBS supernatant and discard.
- Wash the remaining intestinal pieces with 10 ml ice cold PBS, vortex, and remove supernatant with a pipettor and discard.
- Transfer tissue to 5 ml chelation buffer and rock at 4 °C for 10 min.
- Remove chelation buffer from the tissue with a pipettor and wash tissue twice with 10 ml ice cold PBS.
- Add 5 ml ice cold PBS.
- Shake for 2 min. Do not shake with too much vigor. The shake involves full arm motion from the elbow at an even tempo (one down stroke every second, Video 1).Video 1. Crypt isolation example
- Pour off supernatant from first shake and discard because this supernatant will primarily contain villi.
- Add 5 ml PBS and repeat shake for 2 min. Check the supernatant for crypts. To initially check crypts, pipette a 20 μl drop of the PBS supernatant onto a well of a 12 well cell culture dish and view at 4x magnification under a cell culture scope. Look for multiple crypts in which the dark granularity of the Paneth cells can be seen (Figure 2). After this shake, the supernatant should primarily contain villi and look like the image on the left of Figure 2.
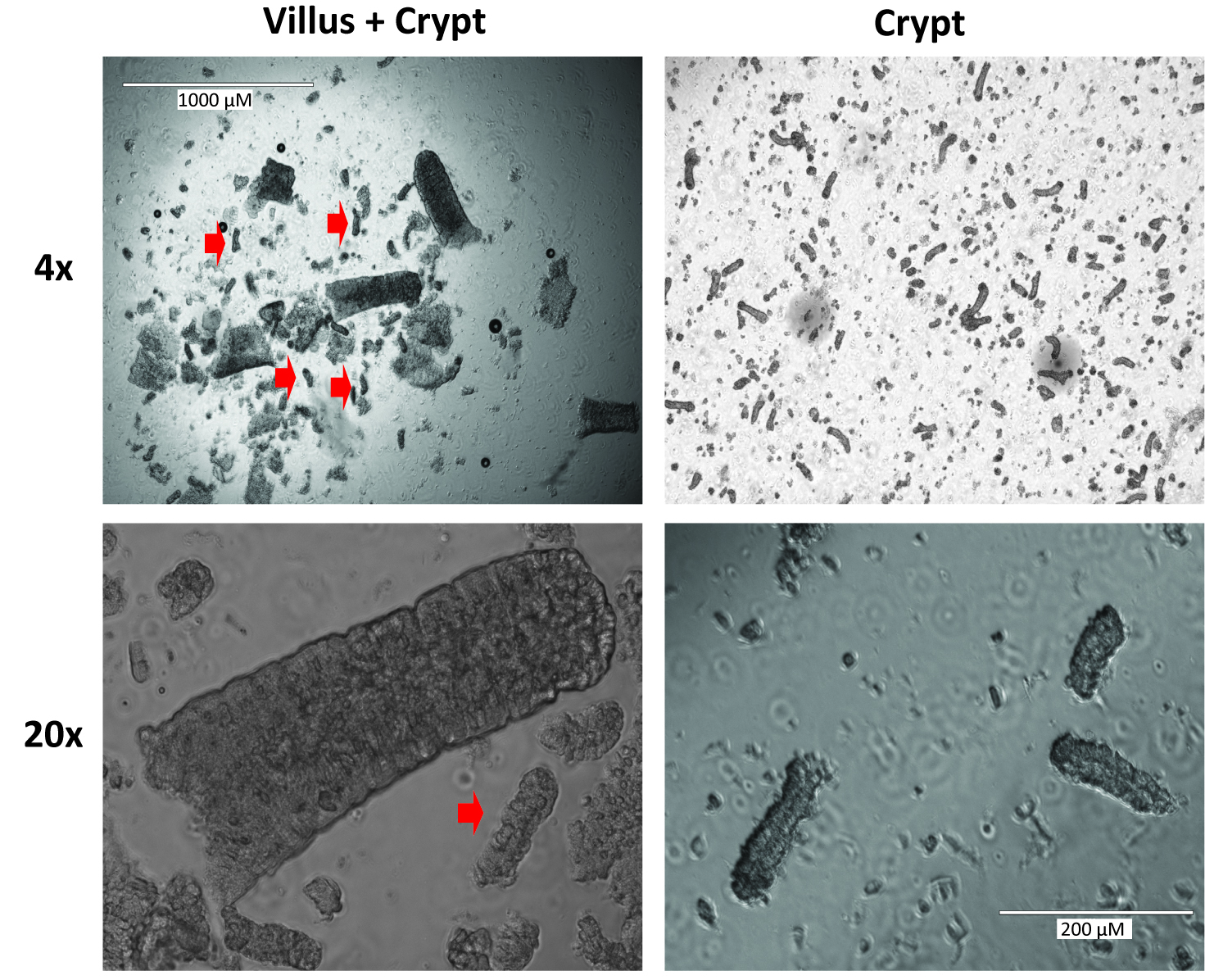
Figure 2. Representative images of villi and crypts at 4x and 20x magnification. Red arrow heads indicate crypts. The images on the left are representative of those likely seen after step A13. The images on the right demonstrate the optimal crypt composition which will be seen after step A18. - Usually, crypts are not seen here and the supernatant can be poured off as well, but this is very dependent on shaking technique so the supernatant should be checked for crypts.
- Add 5 ml fresh chelation buffer and chelate for 10 min at 4 °C on the rocker.
- Decant the chelation buffer, being careful not to lose the 1 cm pieces of small intestine then add 5 ml ice cold PBS (wash). Discard the PBS and perform a second wash. Discard the wash and add 5 ml ice cold PBS for shaking.
- Shake for 2 min. Check for crypts. If you have them here, you can filter and save, but usually the best crypts are seen after a second 2-min shake. Though results will differ based on shaking variability, the supernatant from this shake will usually contain 90% villi and 10% or less crypts.
- Perform a second 2-min shake and check for crypts. After this second shake, there is typically an equivalent number of crypts and villi. Ultimately, a ratio of greater than or equal to 70% crypts to 30% villi is optimal. If crypts are still not seen in abundance, shake more vigorously for 30 sec intervals and continue to check for crypts until at least 20 crypts per 20 μl aliquot are seen.
- Filter through a 70 μm filter into a 50 ml tube. Rinse filter with 5 ml cold shaking buffer.
- Crypts are clearly visible at 4x magnification and can be counted within a representative 20 μl crypt aliquot. At least 20 crypts per 20 μl of the solution should be expected, resulting in a typical total yield of approximately 5,000 crypts. Count crypts and transfer enough volume for 1,500 crypts per two wells to pre-chilled 15 ml conical tubes.
- Centrifuge at 1,000 rpm for 10 min at 4 °C. Aspirate off shaking buffer being careful not to disturb the pellet.
- Using pre-chilled pipette tips and maintaining all reagents on ice, resuspend crypts in 50 μl of Matrigel per well, containing:
- EGF: 50 ng/ml (0.25 μl of 100 μg/ml per 50 μl of Matrigel)
- Noggin: 100 ng/ml (1 μl of 50 mg/ml per 50 μl of Matrigel)
- R-Spondin: 500 ng/ml (1 μl of 250 μg/ml per 50 μl of Matrigel)
- For initial cultures, can also spike with Wnt3a (2 μl 50 μg/ml Wnt3a per 50 μl of Matrigel). This step is not necessary but will increase the plating efficiency of the enteroids.
a. Store growth factors in very small aliquots at -80 °C.
b. When resuspending crypts, be sure to resuspend completely without adding bubbles. Pipette in and out using a P200 set for a volume 25 μl less than the total volume and never fully expel or pull in the Matrigel/crypt mixture. This step can be performed at room temperature in a cell culture hood, keeping reagents in an ice bucket until plating. - EGF: 50 ng/ml (0.25 μl of 100 μg/ml per 50 μl of Matrigel)
- Using pre-chilled pipette tips, plate onto the coverslip portion of the MatTek plate so that the Matrigel forms a mounded dome in the center of the well (Figure 3A).
- Place plate in incubator at 37 °C for 30 min (or longer, up to 2 h) to allow Matrigel to polymerize fully.
- Overlay Matrigel with 350 μl Minigut culture media (Figure 3B).
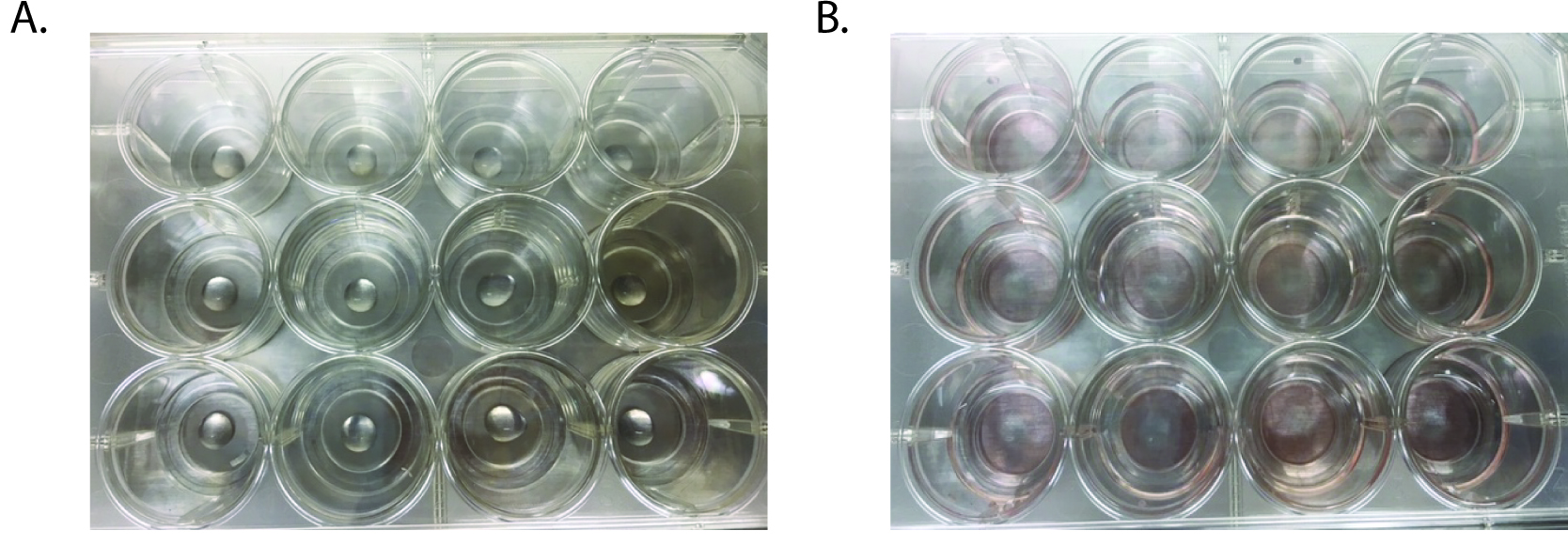
Figure 3. Images of matrigel plating onto 12-well MatTek plates. A. Matrigel containing crypts is pipetted into the center of the well, avoiding any bubble formation. B. After the Matrigel has set, media is added to each well.
- Anesthetize and sacrifice mouse with isofluorane followed by cervical dislocation.
- Proliferation staining
- Proliferation staining can be conducted at any point post-enteroid plating, depending on the process to be analyzed. If initial growth rates are in question, enteroids can be stained after only a day in culture. If response to treatments is to be ascertained, it is best to allow the enteroids to grow for at least 4 days prior to proliferation staining to ensure that they have formed full crypts (see Figure 4 for comparisons of enteroids at different times post-plating). Treatment can also be performed at a specified time prior to staining, depending on the desired output.
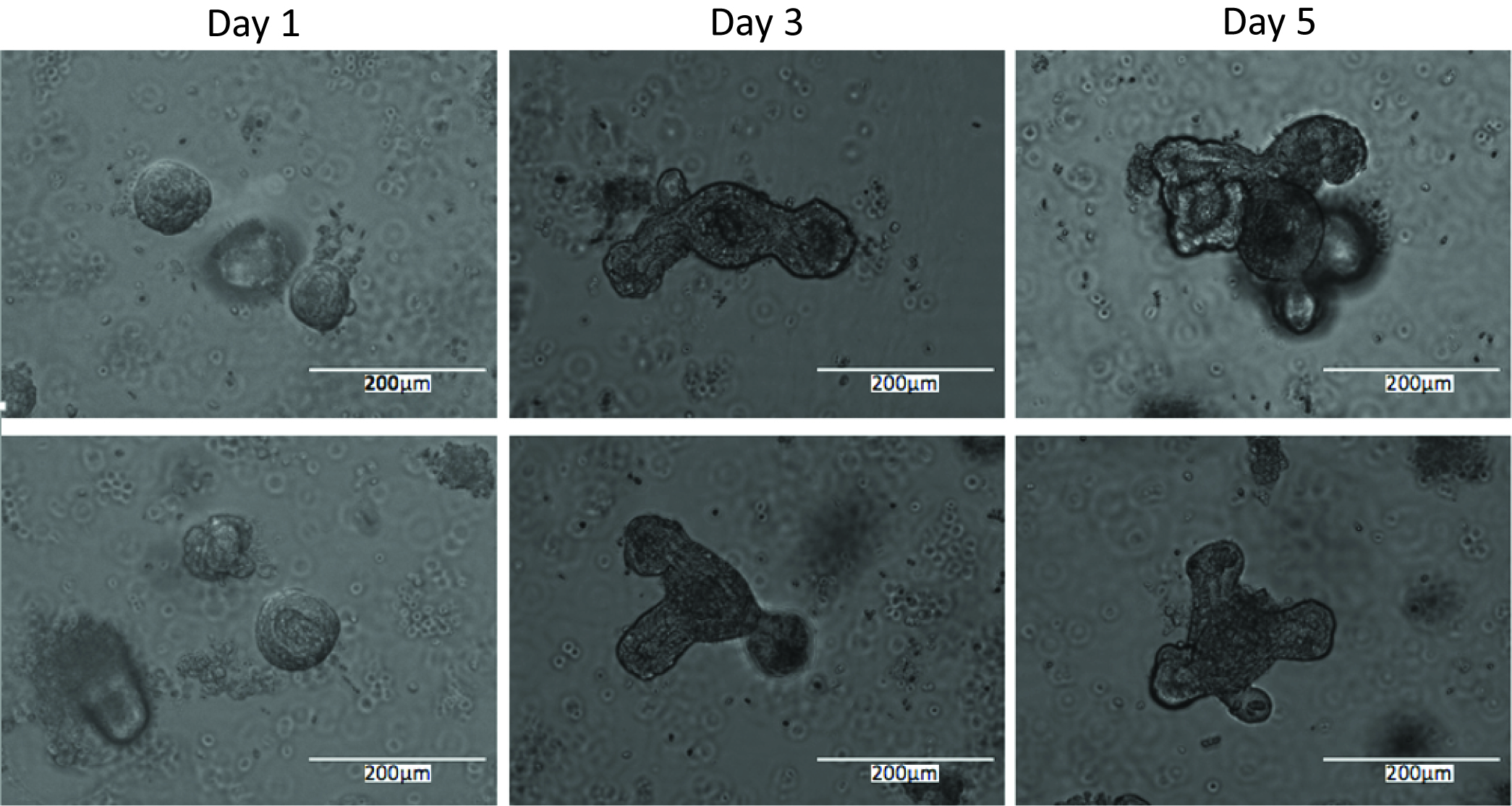
Figure 4. Representative images of enteroids 1 day, 3 days, and 5 days post-plating - Proliferation is determined using the Click-iT EdU cell proliferation assay according to the manufacturer’s instructions as follows.
- Enteroid media is replaced with fresh pre-warmed Minigut media and a 10 μM working solution of EdU.
- Incubate enteroids for 15 min under normal growth conditions.
- Remove media from enteroids and replace with 1 ml of 2% formaldehyde in PBS and incubate overnight at 4 °C.
- The next morning, gently remove the fixative and wash the enteroids twice with 1 ml of 3% BSA in PBS.
- Gently remove the last wash and add 1 ml of 0.5% Triton X-100 in PBS and incubate at room temperature for 20 min.
- Prepare the Click-iT reaction cocktail (according to manufacturer’s instructions) and add 500 μl per well. Rock the plate briefly to ensure even distribution. Incubate the enteroids for 30 min at room temperature, protected from light.
- Remove the reaction cocktail and wash each well once with 1 ml 3% BSA in PBS.
- Remove wash solution and counterstain nuclei with 500 μl of TO-PRO-3 (1:500 in PBS) for 15 min at room temperature. TO-PRO-3 enables both non-proliferating and proliferating cells to be identified by their nuclei so that proliferation percentages can be determined.
- Proliferation staining can be conducted at any point post-enteroid plating, depending on the process to be analyzed. If initial growth rates are in question, enteroids can be stained after only a day in culture. If response to treatments is to be ascertained, it is best to allow the enteroids to grow for at least 4 days prior to proliferation staining to ensure that they have formed full crypts (see Figure 4 for comparisons of enteroids at different times post-plating). Treatment can also be performed at a specified time prior to staining, depending on the desired output.
- Imaging of small intestinal enteroids
- Enteroids can be imaged on an LSM 510 META or similar inverted confocal microscope. Imaging is performed in the 488 (EdU) and 647 (TO-PRO-3) channels and Z-stacks are taken to ensure three-dimensional imaging of the enteroid (Figure 5A-B).
- Proliferation can be quantified as EdU+ cells per crypt area where crypt area is determined using ImageJ image analysis software. Alternatively, proliferation can be quantified as percentage of EdU+ cells relative to total cell number as determined by TO-PRO-3+ nuclei.
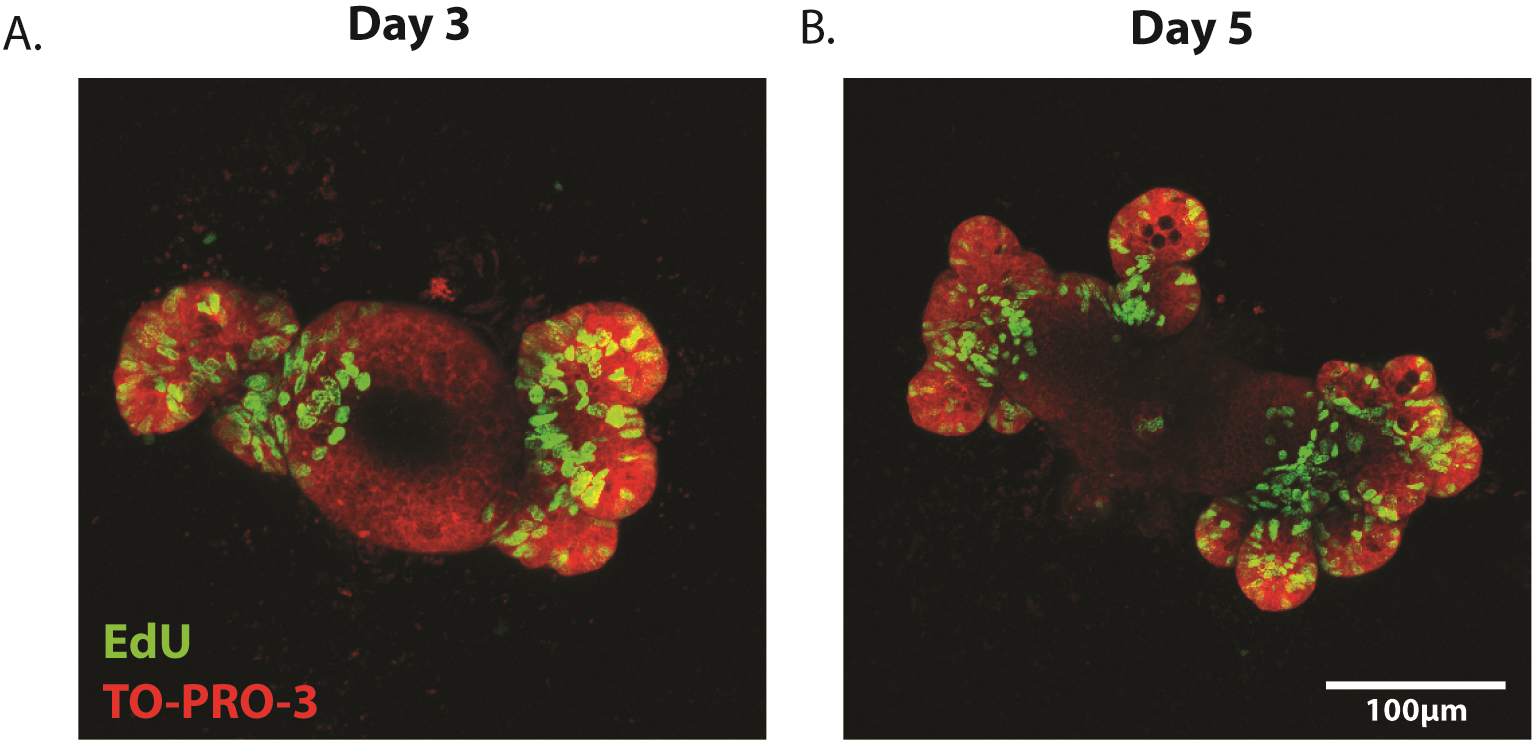
Figure 5. Representative images of proliferating enteroids. Enteroids have been stained for the proliferation marker EdU (green) at (A) 3 days and (B) 5 days post-plating. Counterstain is TO-PRO-3 (nuclei, red).
- Enteroids can be imaged on an LSM 510 META or similar inverted confocal microscope. Imaging is performed in the 488 (EdU) and 647 (TO-PRO-3) channels and Z-stacks are taken to ensure three-dimensional imaging of the enteroid (Figure 5A-B).
Notes
- It is essential to differentiate between crypts and villi (Figure 1). Become comfortable with detecting crypts and determine the optimal shaking protocol for maximal crypt isolation to ensure optimal plating efficiency. Villi are commonly separated from the mucosa in earlier shakes and crypts in later shakes. Perform experiments to increase reproducibility in your hands.
- Ensure that the chelation buffer does not contain calcium (PBS without additional calcium or magnesium) as calcium will disrupt the chelation procedure.
Recipes
- Chelation buffer (prepare fresh)
1 mM EDTA
PBS
Note: Make sure PBS is correct pH. Best to use the purchased 1x DPBS without calcium or magnesium for all solutions. - Shaking buffer
43.3 mM sucrose
54.9 mM sorbitol
PBS
Can be stored at 4 °C - Minigut culture media
Advanced DMEM/F12
2 mM L-glutamine
200 U/ml penicillin
200 μg/ml streptomycin
Freeze this in 43.5 ml aliquots and, upon thawing, add:
20 mM HEPES
1x N2 supplement
1x B27 supplement
10% FBS
Note: If growing cultures for longer than 4 days, every 4 days, media should be replaced with fresh, complete Minigut media with the following growth factors:
1 μg/ml R-spondin 1
100 ng/ml Noggin
50 ng/ml EGF - 2% formaldehyde
2 g paraformaldehyde powder
100 ml of 1x PBS
Heat to, but do not exceed, 70 °C in a fume hood until the paraformaldehyde dissolves
Allow the solution to return to room temperature
Adjust the pH to 7.4 as needed
Filter and stored at 4 °C
Acknowledgments
This protocol was first described in Barrett et al. (2015). This work was supported by NIH grants DK080221 (to C. S. Williams), 1F31CA167920 (to C. W. Barrett), T32CA009592-26 (to C. W. Barrett), and Merit Review Grants from the Office of Medical Research, Department of Veterans Affairs, 1I01BX001426 (to C. S. Williams).
References
- Barrett, C. W., Reddy, V. K., Short, S. P., Motley, A. K., Lintel, M. K., Bradley, A. M., Freeman, T., Vallance, J., Ning, W., Parang, B., Poindexter, S. V., Fingleton, B., Chen, X., Washington, M. K., Wilson, K. T., Shroyer, N. F., Hill, K. E., Burk, R. F. and Williams, C. S. (2015). Selenoprotein P influences colitis-induced tumorigenesis by mediating stemness and oxidative damage. J Clin Invest 125(7): 2646-2660.
- Mahe, M. M., Aihara, E., Schumacher, M. A., Zavros, Y., Montrose, M. H., Helmrath, M. A., Sato, T. and Shroyer, N. F. (2013). Establishment of gastrointestinal epithelial organoids. Curr Protoc Mouse Biol 3(4): 217-240.
- Sato, T., Vries, R. G., Snippert, H. J., van de Wetering, M., Barker, N., Stange, D. E., van Es, J. H., Abo, A., Kujala, P., Peters, P. J. and Clevers, H. (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459(7244): 262-265.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Barrett, C. W., Short, S. P., Choksi, Y. A. and Williams, C. S. (2016). Whole-mount Enteroid Proliferation Staining. Bio-protocol 6(12): e1837. DOI: 10.21769/BioProtoc.1837.
Category
Immunology > Immune cell isolation > Maintenance and differentiation
Immunology > Immune cell function > General
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










