- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Differential and Simultaneous Visualization of Cells and Airspaces in Plant Leaves
Published: Vol 6, Iss 11, Jun 5, 2016 DOI: 10.21769/BioProtoc.1826 Views: 8698
Reviewed by: Tie LiuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Near-Infrared Autofluorescence Imaging of Nuclei in Living Plant Roots
Akira Yoshinari and Masayoshi Nakamura
Apr 20, 2025 2132 Views

ClearDepth Method for Evaluations of Root Depth in Soil-Filled Pots
Michel Ruiz Rosquete [...] Wolfgang Busch
Aug 20, 2025 2129 Views

Live-Cell Monitoring of Piecemeal Chloroplast Autophagy
Masanori Izumi [...] Shinya Hagihara
Nov 5, 2025 1690 Views
Abstract
This protocol is to differentially and simultaneously visualize both cells and airspaces in intact leaves and to create 3D structures of cells and airspaces from confocal images using the software DSLT_Demo (https://github.com/nslab2000/DSLT). Leaves stained with Nile Red in silicone-oil solution provide red color to cell membranes and green color to airspaces filled with silicone oil solution. This method is applicable to any tissues (except for dry seeds) of various plants including Arabidopsis, Nicotiana, Lemna and moss, and applicable even to hard leaves of plants such as switchgrass and Cinnamomum. Repeated use of this method enables time-lapse imaging of leaves over days and weeks because both Nile Red and silicone oil are harmless to plant tissues.
Materials and Reagents
- >100 ml container to hold the staining solution (see procedure A) during rotational incubation
- An incubation container for staining (e.g., 1.5 ml micro tube, Petri dish)
- Super frost slide glass (Matsunami Glass Ind., catalog number: S2441 )
- NEO microscope coverslip, 24 x 32 mm (Matsunami Glass, catalog number: C024321 )
- Parafilm M® (Bemis Company, Inc., catalog number: PM-996 )
- Pipetman p200 or 1 ml disposable transfer pipette
- Silicone oil (Shin-Etsu Chemical Co., catalog number: KF96L-1.0 cs or KF96L-1.5 cs )
Note: It is named “Dimethyl Silicone Fluid” on Shin-Etsu Chemical Co. website. - Nile Red (Invitrogen, catalog number: N-1142 )
Note: Currently, it is “Thermo Fisher Scientific, Molecular Probes™, catalog number: N-1142”.
Equipment
- Tube rotator or shaker
- Electronic scale
- Confocal microscope (Carl Zeiss Microscopy, model: LSM-780) or fluorescence microscope (Carl Zeiss Microscopy, model: Axioskop2 plus)
- Sonication bath (HONDA ELECTRONICS Co., model: W-113 )
Note: This is not absolutely necessary.
Procedure
- Preparation of staining solution
- Add 0.318 mg of Nile Red and 100 ml of silicone oil (KF96L-1.0 cs or KF96L-1.5 cs) to a container. The final concentration of this solution is 10 μM. About 100 samples can be prepared from this amount of staining solution if you use 1.5 ml microtubes for staining.
- Tightly seal the container with Parafilm M®.
- (Optional) Sonicate the suspended Nile Red using a sonication bath (10 min, 100 kHz) to make Nile Red dissolved faster.
- Wrap the container with aluminum foil, place the tube on a tube rotator and rotate for 1-2 d.
- (Optional) Undissolved dye can be removed via filtration or centrifugation.
- Add 0.318 mg of Nile Red and 100 ml of silicone oil (KF96L-1.0 cs or KF96L-1.5 cs) to a container. The final concentration of this solution is 10 μM. About 100 samples can be prepared from this amount of staining solution if you use 1.5 ml microtubes for staining.
- Staining
- Remove a small (5 x 5 ~ 10 x 10 mm) section of the leaf tissue to be examined.
Note: When you conduct time-lapse imaging, skip this step and stain a whole plant. - Add the specimen and staining solution to an incubation container.
- Incubate specimen in staining solution for 5 min (adaxial side) or 15 min (abaxial side).
- Remove a small (5 x 5 ~ 10 x 10 mm) section of the leaf tissue to be examined.
- Slide preparation
Prepare slides according to Figure 1.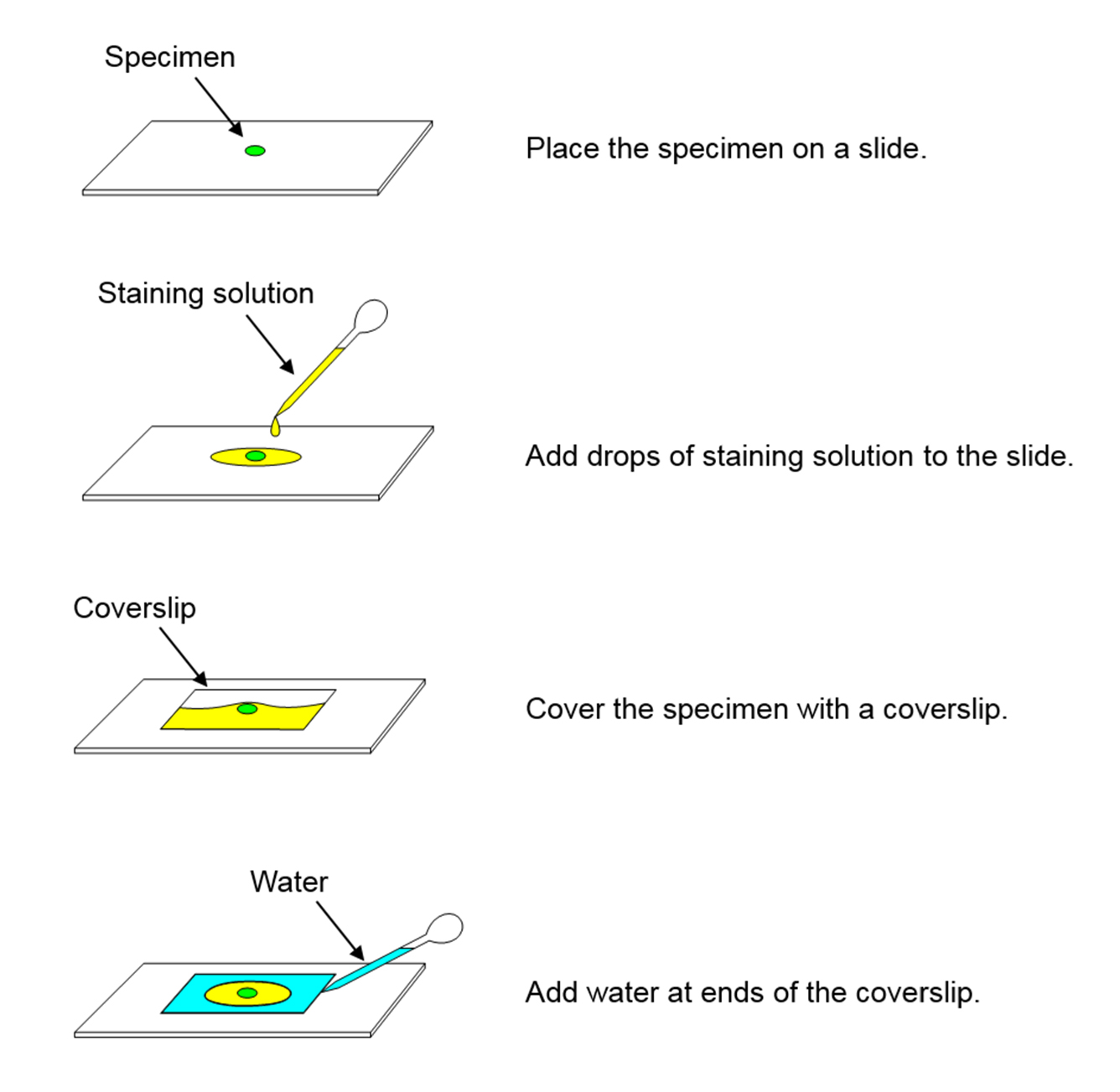
Figure 1. Procedure of slide preparation - Microscopy
The steps B-C fill airspaces with Nile Red in silicone oil. Image airspaces (excitation wavelength 488, emission wavelength 490-560) and cells (excitation wavelength 561, emission wavelength 563-639). Representative data is shown in Figure 2. - (Optional) Procedures for time-lapse imaging
After observation, shake the plants in silicone oil (KF96L-1.0 cs) gently at 30~60 rpm for 1 min twice to remove Nile Red. Put the plants on culture beds until the next observation. Repeat the steps B-E. Representative data of time-lapse imaging is shown in Figure 3. - Image segmentation
You can extract shapes of cells and airspaces from acquired 3D images by using DSLT_Demo. Step by step instructions for segmentation are available at http://dslt.bot.kyoto-u.ac.jp/DSLT_manual_v111.pdf.
Representative data
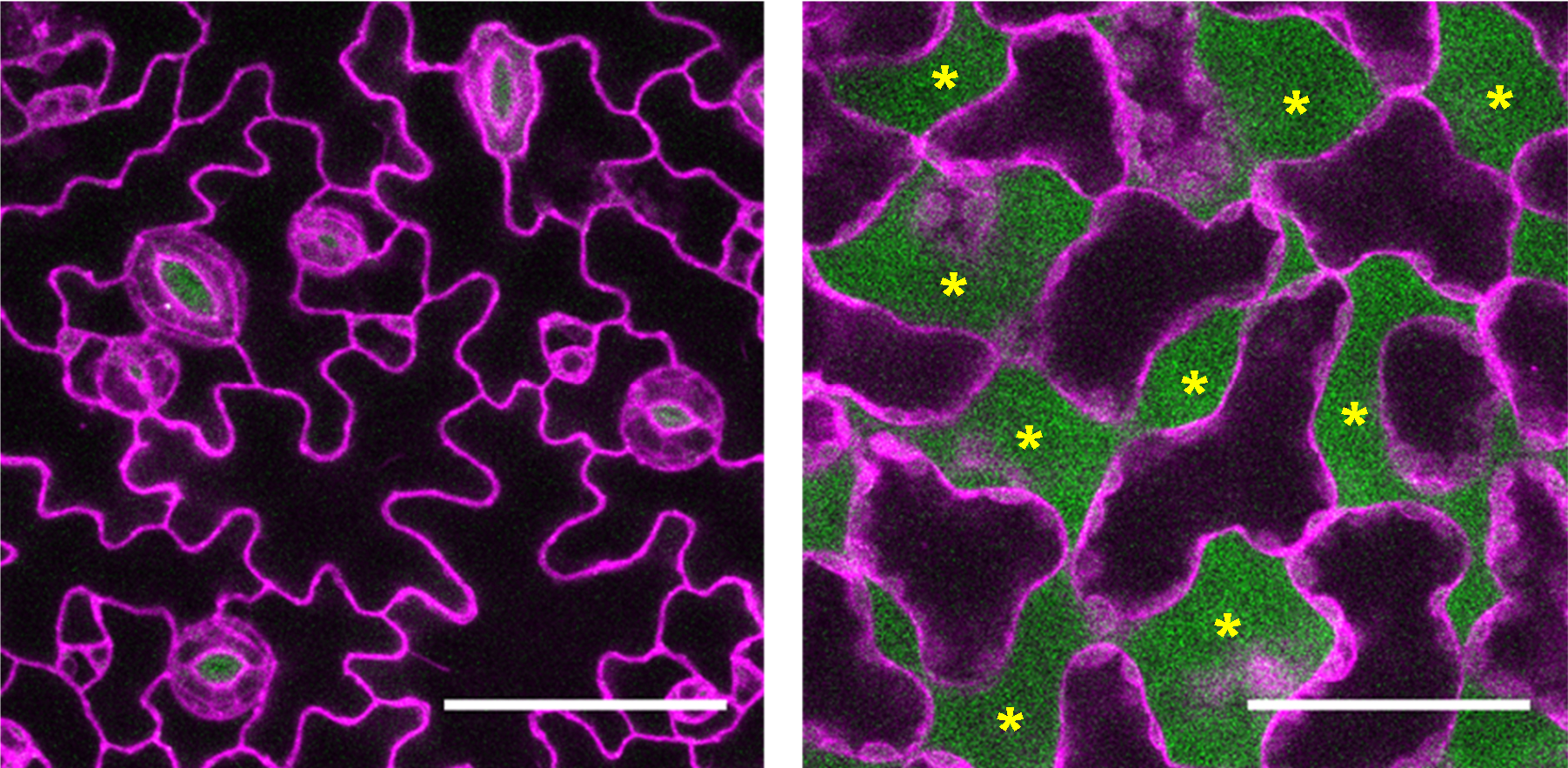
Figure 2. Visualizing cells and airspaces in an Arabidopsis thaliana leaf. Confocal images of the epidermis (left) and mesophyll (right) of an Arabidopsis thaliana leaf. The abaxial side of the leaf was stained with solution of Nile Red in silicone oil and imaged with 488 nm excitation for Nile Red in silicone oil (airspace, green) and 561 nm excitation for Nile Red in lipid (cellular membrane, magenta). Asterisks indicate airspaces. Scale bars: 50 μm.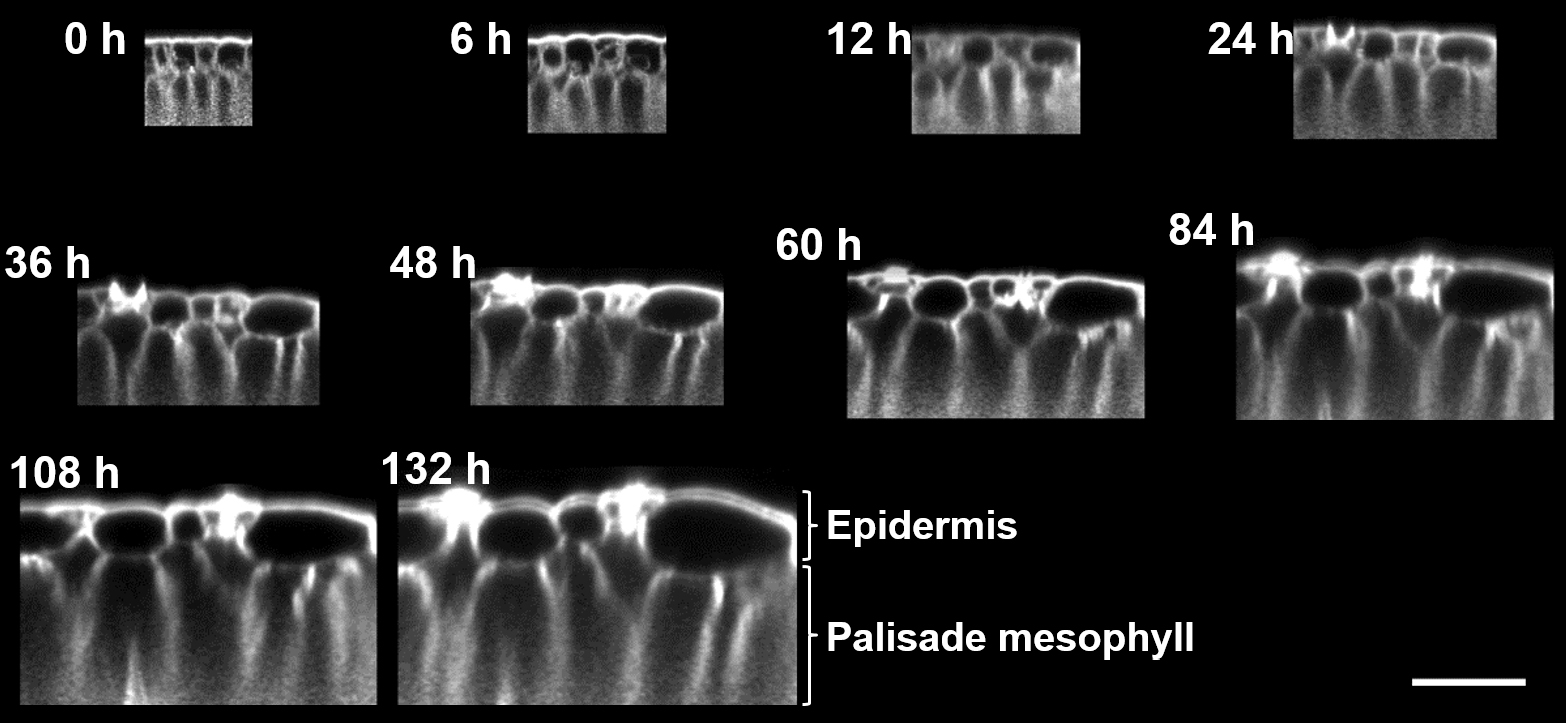
Figure 3. Time-lapse imaging of a developing Nicotiana glauca leaf. Cross-sectional views of 3D images of the adaxial side of a Nicotiana glauca leaf. Confocal images were taken over 5 days of leaf development (times indicated in white above each image). Shown is the fluorescence of Nile Red in the lipid bilayer membrane. Bar is 50 μm.
Acknowledgments
This protocol was adapted from the research article Kawase et al. (2015). The work was supported by a Specially Promoted Research of Grant-in-Aid for Scientific Research to I. H. -N. (no.22000014) from the Japan Society for the Promotion of Science (JSPS).
References
- Kawase, T., Sugano, S. S., Shimada, T. and Hara-Nishimura, I. (2015). A direction-selective local-thresholding method, DSLT, in combination with a dye-based method for automated three-dimensional segmentation of cells and airspaces in developing leaves. Plant J 81(2): 357-366.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Kawase, T., Sugano, S. S., Shimada, T. and Hara-Nishimura, I. (2016). Differential and Simultaneous Visualization of Cells and Airspaces in Plant Leaves. Bio-protocol 6(11): e1826. DOI: 10.21769/BioProtoc.1826.
Category
Plant Science > Plant physiology > Phenotyping
Cell Biology > Cell imaging > Confocal microscopy
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









