- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Nitrite Reduction Assay for Whole Pseudomonas Cells
Published: Vol 6, Iss 10, May 20, 2016 DOI: 10.21769/BioProtoc.1818 Views: 12232
Reviewed by: Valentine V TrotterGeneviève BallAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

H2 Production from Methyl Viologen–Dependent Hydrogenase Activity Monitored by Gas Chromatography
Nuttavut Kosem
Dec 5, 2023 1769 Views

Monitoring Protein Stability In Vivo Using an Intein-Based Biosensor
John S. Smetana [...] Christopher W. Lennon
Apr 20, 2025 1584 Views

Endo-1,4-β-D-xylanase Assay Using Azo-Xylan and Variants Thereof
Luca Bombardi [...] Salvatore Fusco
Apr 20, 2025 1927 Views
Abstract
The second step of the dissimilatory denitrification pathway in which nitrite (NO2-) is converted to nitric oxide (NO) is catalyzed by the enzyme nitrite reductase. Two distinct enzymes are found in nature that catalyze this reaction, and they contain different metal sites, either iron (Fe), in the form of heme, or copper (Cu) (Zumft, 1997). The Pseudomonas stutzeri (P. stutzeri) RCH2 strain used in this assay contains both an Fe and a Cu form of nitrite reductase. In this assay, total nitrite reductase activity can be measured in whole cells using fumarate or some other carbon source as an electron source by measuring the disappearance of nitrite over time (Thorgersen et al., 2015).
Keywords: Nitrite reductaseMaterials and Reagents
- Borosilicate glass culture tubes, 16 x 125 mm (VWR International, catalog number: 47729-578 )
- Hungate tubes (16 x 125 mm) with butyl rubber stopper (Bellco Glass, catalog number: 2047-16125 )
- Pseudomonas cells
- M9, minimal salts, 5x (Sigma-Aldrich, catalog number: M6030 )
- Bacto yeast extract technical (BD Biosciences, catalog number: 288610 )
- Potassium phosphate (dibasic, powder) (VWR International, J.T. Baker®, catalog number: 3252-05 )
- Potassium phosphate (monobasic, crystal) (VWR International, J.T. Baker®, catalog number: 3246-05 )
- Sodium fumarate dibasic (Sigma-Aldrich, catalog number: F1506 )
- Sodium nitrate (VWR International, J.T. Baker®, catalog number: 3770-01 )
- Sodium nitrite (VWR International, J.T. Baker®, catalog number: 3780-01 )
- Griess reagent (Sigma-Aldrich, catalog number: G4410 )
- High purity 100% argon gas (Airgas, catalog number: ARHP300 )
- 50 mM potassium phosphate buffer (pH 7.0) (see Recipes)
- Assay buffer (see Recipes)
Equipment
- Allegra 25R centrifuge (Beckman Coulter)
- Spectrophotometer (measuring absorption in the visible range)
- Innova 4230 refrigerated incubator shaker (New Brunswick Scientific)
Procedure
- Cultures (5 ml) of P. stutzeri RCH2 were grown anaerobically with shaking (250 rpm) at 30 °C in Hungate tubes on M9 salts supplemented with 20 mM fumarate as a carbon source, 20 mM nitrate as an electron acceptor, and 0.5 g/L yeast extract. Cells were harvested during late log phase (OD650 0.7-0.9) by centrifugation at 4 °C (10 min at 5,000 x g) and washed once with pre-chilled (4 °C) 50 mM potassium phosphate buffer (pH 7.0) before being resuspended in the same buffer at approximately 3-7 x 109 cells/ml.
- In a culture tube, 500 μl of cell suspension was added to 4.5 ml of assay buffer. The assay was incubated at 30 °C with slow shaking (150 rpm) in an incubator. The volume of cells added can be changed to optimize reaction speed.
- Samples (100 μl) were taken every 5 min or longer as needed for 20 min to1 h, and were diluted into 900 μl distilled water. One ml of Griess reagent was added to the samples and they were incubated for 15 min at room temperature.
- The visible absorption at 540 nm was measured for each sample, and nitrite concentrations were calculated using a nitrite standard curve made in 50 mM potassium phosphate buffer (pH 7.0) (0-100 μM nitrite) (Figure 1). A unit of nitrite reductase activity catalyzed the reduction of 1 nmol of nitrite/min. A negative control using 50 mM potassium phosphate buffer (pH 7.0) without cells was also performed.
Representative data
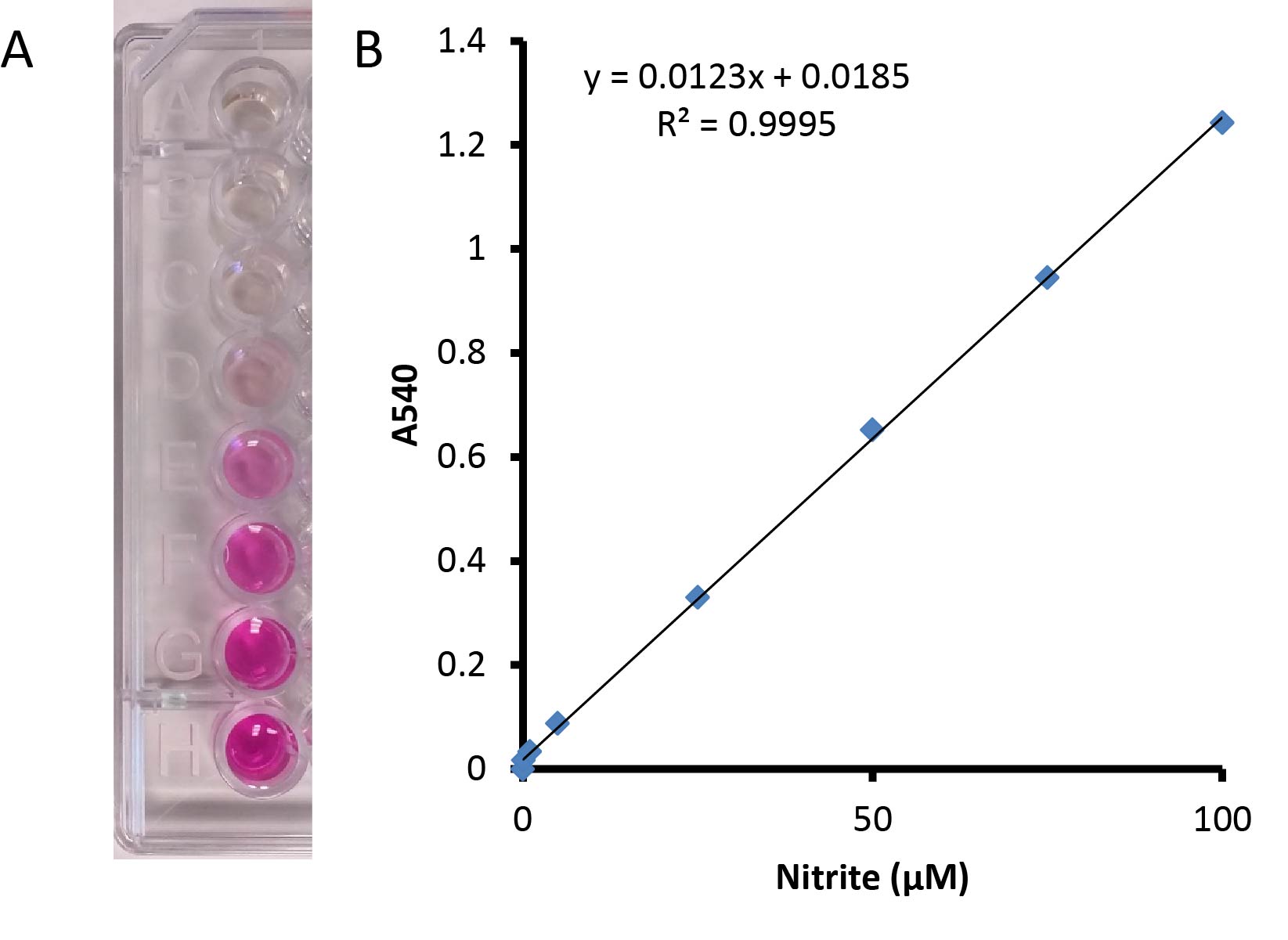
Figure 1. Standard nitrite curve from 0-100 µM nitrite mixed 1:1 with Griess reagent. A. The red-pink color develops when the Griess reagent is added to the nitrite containing sample. B. A linear standard curve is shown with values ranging from 0-100 µM nitrite.
Notes
- If there is a concern about oxygen lability of the nitrite reductase enzyme, the method can optionally be performed anoxically with the use of sealed Hungate tubes where the headspace is exchanged with high purity 100% argon. The nitrite reductase activity of P. stutzeri RCH2 was stable in the presence of oxygen over the time period of the assay.
- Other carbon sources other than fumarate can be used to fit the carbon source preferences of the microorganism being studied including but not limited to lactate, pyruvate, and glucose. Nitrite reductase may not be as highly expressed if the microorganism can ferment the carbon source used rather than depending on nitrate or nitrite as electron acceptors. Additionally, cells were grown anaerobically in the presence of nitrate as an electron acceptor to increase nitrite reductase expression. Nitrite (2-10 mM) could also be used as an electron acceptor depending on the tolerance of the organism being assayed.
Recipes
- 50 mM potassium phosphate buffer (pH 7.0)
- 1 M solutions of KH2PO4 (A) and K2HPO4 (B) were made separately.
- A mixture containing 39% (v/v) solution A and 61% (v/v) solution B was made.
- The pH of the mixture was adjusted to pH 7.0 with solution A or B before diluting.
- Dilute the stock solution 1:20 with distilled water (final concentration: 50 mM).
- 1 M solutions of KH2PO4 (A) and K2HPO4 (B) were made separately.
- Assay buffer
50 mM potassium phosphate buffer (pH 7.0)
40 mM fumarate
1 mM nitrite (pH 7.0)
Acknowledgments
This material by ENIGMA (Ecosystems and Networks Integrated with Genes and Molecular Assemblies) (http://enigma.lbl.gov), a Scientific Focus Area Program at Lawrence Berkeley National Laboratory, is based upon work supported by the U. S. Department of Energy, Office of Science, Office of Biological and Environmental Research, under contract number DE-AC02-05CH11231.
References
- Thorgersen, M. P., Lancaster, W. A., Vaccaro, B. J., Poole, F. L., Rocha, A. M., Mehlhorn, T., Pettenato, A., Ray, J., Waters, R. J., Melnyk, R. A., Chakraborty, R., Hazen, T. C., Deutschbauer, A. M., Arkin, A. P. and Adams, M. W. (2015). Molybdenum availability is key to nitrate removal in contaminated groundwater environments. Appl Environ Microbiol 81(15): 4976-4983.
- Zumft, W. G. (1997). Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61(4): 533-616.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Thorgersen, M. P. and Adams, M. W. W. (2016). Nitrite Reduction Assay for Whole Pseudomonas Cells. Bio-protocol 6(10): e1818. DOI: 10.21769/BioProtoc.1818.
Category
Microbiology > Microbial biochemistry > Protein > Activity
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










