- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro Deneddylation Assay
Published: Vol 6, Iss 6, Mar 20, 2016 DOI: 10.21769/BioProtoc.1756 Views: 10109
Reviewed by: Valentine V TrotterManuela RoggianiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Measurement of Energy-dependent Rhodamine 6G Efflux in Yeast Species
Yvetta Gbelska [...] Alexandra Konecna
Aug 5, 2017 8961 Views

An Assay to Determine NAD(P)H: Quinone Oxidoreductase Activity in Cell Extracts from Candida glabrata
Anamika Battu [...] Rupinder Kaur
Nov 5, 2021 3061 Views

Isolation of Mitochondria from Ustilago maydis Protoplasts
Juan Pablo Pardo [...] Lucero Romero-Aguilar
Jan 5, 2022 3581 Views
Abstract
Nedd8 is a small ubiquitin-like protein (9 kDa) covalently attached to a conserved lysine residue of a cullin protein which is part of cullin-RING ligases (CRLs). CRLs are major E3 ligases important for protein ubiquitination in the ubiquitin-proteasome pathway (UPP). The activity of CRLs is regulated by cycles of neddylation (CulA-N8, ~98 kDa) and deneddylation (CulA ~89 kDa). The COP9 signalosome (CSN) and Deneddylase A (DenA) are capable of cleaving the isopeptide bond between Nedd8 and CullinA. In contrast to the single protein DenA, CSN is an eight subunit multiprotein complex. Protein crude extracts of different Aspergillus nidulans csn deletion strains were mixed with recombinant CSN subunits expressed and purified from Escherichia coli (E. coli). Western hybridization experiments using anti-CulA or anti-Nedd8 antibodies could show the ratio of neddylated vs. deneddylated CulA. Using the deneddylation assay, we could show that CsnE is the last subunit joining a 7-subunit pre-assembled CSN in vitro and only then CSN can perform cullin deneddylation by the metalloprotease subunit CsnE. This assay is a fast and non-expensive method, which visualizes enzyme activity for deneddylating proteins. It might be also useful for testing the activity of other isopeptidases removing post-translational modifications from substrates in Aspergillus nidulans (A. nidulans) or other organisms.
Keywords: DeneddylationMaterials and Reagents
- Recombinant protein overexpression and protein purification
- 1 ml cuvettes 10 x 10 x 45 mm (Sarstedt AG, catalog number: 67.742 )
- Chromatography columns [e.g. GSTrap (GE Healthcare, catalog number: 17-5130-01 ), HisTrap (GE Healthcare, catalog number: 17-5255-01 ), Superdex 200 HR 10/300 GL (GE Healthcare, catalog number: 17-5175-01 ), HiPrep 16/60 Sephacryl S-300 HR (GE Healthcare, catalog number: 17-1167-01 )]
- Amicon Ultra centrifugal filters (Merck Millipore Corporation) molecular weight cut off (MWCO) depends on protein size [e.g. 30 kDa (Merck Millipore Corporation, MWCO, catalog number: UFC903024 )]
- MicronSep filter paper, 0.22, 47 mm (Carl Roth GmbH + Co., catalog number: A009.1 )
- Escherichia coli (E. coli) strains suitable for protein overexpression (e.g. Rosetta DE3) transformed with overexpression plasmid including protein sequence of interest with N- or C-terminal Glutathione-S-transferase- (GST-) or Histidine- (His-) tag
- Tryptone/Peptone from Casein (Carl Roth GmbH + Co., catalog number: 8952.2 ) used as 1% solution
- Yeast extract (Oxoid, catalog number: LP0021 ) used as 0.5% solution
- Sodiumchloride (NaCl) (Carl Roth GmbH + Co., catalog number: 9265.2 ) used as 1% solution
- Antibiotics according to the E. coli strains used
- Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Carl Roth GmbH + Co., catalog number: 2316.4 ) used as 1 M solution, filter sterilized
- 99.5% denatured Ethanol with 1% MEK (VWR International, catalog number: 85033.460 ) used as 20% solution diluted in ddH2O, filter degassed
- Imidazole
- Glutathione
- Lysogeny broth (LB) medium (see Recipes)
- Lysis buffer (see Recipes)
- Affinity buffer (see Recipes)
- Elution buffer for HisTrap (see Recipes)
- Elution buffer for GSTrap (see Recipes)
- Size exclusion buffer (SEC) (see Recipes)
- Preparation of protein crude extracts
- Paper towels
- Spores of Aspergillus nidulans strains
- Miracloth (Merck Millipore Corporation, catalog number: 475855-1R )
- Sodiumchloride (NaCl) (Carl Roth GmbH + Co., catalog number: 9265.2) used as 0.95 % solution
- Liquid nitrogen (Air Liquid)
- Minimal medium (see Recipes)
- 50x AspA (see Recipes)
- Trace elements (see Recipes)
- B* buffer (see Recipes)
- Paper towels
- Deneddylation assay
- Gel blotting paper (Schleicher & Schuell BioScience GmbH, catalog number: 10426694 )
- Blotting membrane AmershamTM ProtranTM 0.45 µm NC (GE Healthcare, catalog number: 10600002 )
- Methanol (VWR International, catalog number: 20864.320 )
- 2-Propanol HiPerSolv CHROMANORM (VWR International, catalog number: 20880.320 )
- Page ruler prestained protein ladder (Thermo Fisher Scientific, catalog number: 26616 )
- Acrylamid Rotiphorese® Gel 30 (37, 5:1) (Carl Roth GmbH + Co., catalog number: 3029.1 )
- Sucofin skimmed milk powder (TSI GmbH, catalog number: ST00227 )
- α-CulA, α-Nedd8 antibodies (GenScript)
- Monoclonal α-Tubulin antibody (Sigma-Aldrich, catalog number: T0926 )
- α-Actin antibody (Novus Biologicals, catalog number: NB100-74340 )
- α-goat rabbit secondary antibody (Thermo Fisher Scientific, InvitrogenTM, catalog number: G21234 )
- α-goat mouse secondary antibody (Jackson ImmunoResearch, catalog number: 115-035-003 )
- p-coumaric acid (Sigma-Aldrich, catalog number: C9008-5 G ) used as 400 mM stock solution dissolved in DMSO (protect from light, store at -20 °C)
- Luminol (Carl Roth GmbH + Co., catalog number: 4203.1 ) used as 250 mM stock solution dissolved in DMSO (protect from light, store at -20 °C)
- 2-mercaptoethanol (Sigma-Aldrich, Fluka Chemika, catalog number: 63700 )
- Sodium dodecyl sulfate (SDS) (Carl Roth GmbH + Co., catalog number: CN30.3 ) used as 7%, 0.1%, 1%, 0.2% solutions
- Bromophenol blue (Sigma-Aldrich, Riedel-de Haën, catalog number: 32768 ) used as 0.3% solution
- Ammoniumpersulfate (APS) (Sigma-Aldrich, Fluka Analytical, catalog number: 09913-100 G ) used as 10% solution
- Tetramethylethylendiamine (TEMED) (Carl Roth GmbH + Co., catalog number: 2367.1 )
- Tween-20 (Carl Roth GmbH + Co., catalog number: 9127.1 )
- Tris-PUFFERAN® (Carl Roth GmbH + Co., catalog number: 4855.3 ) used as 250 mM solution
- Glycine (Carl Roth GmbH + Co., catalog number: 3187.3 ) used as 2.5 M solution
- 30% Hydrogen peroxide (H2O2) (Merck Millipore Corporation, catalog number: 822287.2500 )
- 3x SDS sample buffer (see Recipes)
- 12% SDS gels (see Recipes)
- 2x resolving buffer (see Recipes)
- 2x stacking buffer (see Recipes)
- 10x SDS running buffer (see Recipes)
- 10x transfer buffer (see Recipes)
- 10x TBST (Tris-Buffered Saline Tween 20) buffer (see Recipes)
- Blocking solution (see Recipes)
- Detection solution A (see Recipes)
- Detection solution B (see Recipes)
- Gel blotting paper (Schleicher & Schuell BioScience GmbH, catalog number: 10426694 )
- Others (material and reagents used for more than one method)
- Micro tube 1.5 ml (Sarstedt AG, catalog number: 72.690.001 )
- Micro tube 2.0 ml (Sarstedt AG, catalog number: 72.691 )
- Tube 115 x 28 mm, PP (Sarstedt AG, catalog number: 62.559.001 )
- 5 ml test tube
- 2 L flasks
- PPCO centrifuge bottles (Slaughte, Nalgene, catalog number: 525-2316 )
- Oak-Ridge centrifugation tubes, 50 ml (Carl Roth GmbH + Co., catalog number: PK03.1 )
- 300 ml baffled flasks
- Funnels
- Crushed ice
- 1, 4- Dithiothreitol (DTT) (Carl Roth GmbH + Co., catalog number: 6908.2 ) used as 1 M solution
- trans-Epoxysuccinyl-L-leucylamido(4-guanidino)butane, E-64 protease inhibitor (Sigma-Aldrich, catalog number: E3132 ) used as 1 M stock solution
- Phenylmethylsulfonyl fluoride (PMSF) (Carl Roth GmbH + Co., catalog number: 6367.3 ) used as 100 mM solution in Isopropanol
- Double destilled water (ddH2O)
- Millipore water
- Sodium chloride (NaCl) (Carl Roth GmbH + Co., catalog number: 9265.2) used as 2 M solution
- Tris- hydrochloride (Tris-HCl) (Carl Roth GmbH + Co., catalog number: 9090.3 )
Note: Prepare a series of Tris-HCl solution with different concentrations and pH including 1 M solutions with pH 6.8, 7.5, 8.0 and pH 8.8; 0.75 M solution withpH 8.8; 100 mM solution with pH 8.0. - Ethylenediaminetetraacetic acid (EDTA) (Carl Roth GmbH + Co.) used as 0.5 mM solution
- Nonoxinol-40 (NP-40) (Affymetrix, USB Corporation, catalog number: 19628 )
- Glycerol (Carl Roth GmbH + Co., catalog number: 3783.1 )
- Complete protease inhibitor cocktail (Roche Diagnostics, catalog number: 11873580001 ) (see Recipes)
- Micro tube 1.5 ml (Sarstedt AG, catalog number: 72.690.001 )
Equipment
- Biowave DNA Llife Science Spectrophotometer (Biochrom WPA, catalog number: 80-3004-70 )
- Centrifuge Sorvall RC3B Plus (Thermo Fisher Scientific)
- Centrifuge Sorvall RC5B Plus (Thermo Fisher Scientific)
- Rotor Sorvall SS34 (Thermo Fisher Scientific)
- ÄKTAexplorer 10 (GE Healthcare, catalog number: 18-1300-00 )
- pH meter pH526 (Sigma-Aldrich, model: WTW )
- Filter and degassing system (Satorius)
- Vacuum pump (KNF NEUBERGER, catalog number: 2.03360238 )
- Sonicator Sonoplus HD2070 (Bandelin)
- Superloop 1/16’’ (GE Healthcare, catalog number: 18-1113-82 )
- Sample loop 5 ml (GE Healthcare, catalog number: 18-1140-53 )
- Mixer Mill MM 400 (Retsch)
- Vortex Genie 2 (Scientific Industries)
- Pipettes (1-5,000 µl) (Gilson)
- Centrifuge Biofuge fresco (Heraeus Instruments GmbH)
- Nanodrop Spectrophotometer (VWR International, Peqlab)
- Mini-PROTEAN® Tetra Cell Casting Module (Bio-Rad Laboratories, catalog number: 1658008 )
- Buffer Tank and Lid (Bio-Rad Laboratories, catalog number: 1658040 )
- Mini Trans-Blot Module (Bio-Rad Laboratories, catalog number: 1703935 )
- Autoradiography casette
- FUSION SL-4 3500.WL (Chemiluminescence System) (PEQLAB)
- CCD camera FUSION 4 MP (VWR International, Peqlab)
- Orbital Shaker 3015 (GFL)
- Shaker (INFORS AG)
- Heating block (Gebr. Liebisch GmbH & Co. KG Labortechnik)
Software
- Fusion BIO-1D software
Procedure
Note: The preparation of buffers, solutions and SDS gels is explained in Recipes section.
- Recombinant protein overexpression and protein purification
- For overexpression of Aspergillus nidulans proteins in Escherichia coli (here: Rosetta DE3) inoculate 5 ml LB pre-culture, supplemented with appropriate antibiotics, for overnight incubation at 37 °C with agitation.
- The next morning, use 1 ml of the pre-culture to inoculate 1 L LB in a 2 L flask supplemented with appropriate antibiotics. Incubate the culture at 37 °C with ~220 rpm agitation for 2 to 3 h until OD600 of 0.6-0.7 is reached (depends on E. coli strain). OD600 can be determined with UV Spectrophotometer by taking 1 ml aliquot of the culture measured against a LB blank sample.
- When OD600 of 0.6-0.7 is reached, add 1 ml 1 M IPTG to the culture to induce the overexpression. Transfer the flask to room temperature or 16 °C for no longer than 20 h and 150-180 rpm agitation.
- Transfer the culture to 1 L centrifugation bottles, which should be pre-cooled on ice and then centrifuge at 4,000 rpm for 30 min at 4 °C.
- Discard the supernatant and keep the pellet on ice. Resuspend the pellet with 5-10 ml ice-cold affinity buffer.
- Transfer the resuspended pellet to a 50 ml tube and centrifuge at 4,000 rpm, 20 min, 4 °C.
- Discard the supernatant and store the pellet at -80 °C.
- For protein purification thaw the pellet on ice while preparing the lysis, affinity and elution buffers. Filter-degas the buffers using the filter and degassing system with MicronSep filters.
- Mix the pellet with 10 ml ice-cold lysis buffer per g pellet and resuspend by pipetting up and down (prevent air bubbles and keep solution on ice).
- Put the falcon in a box or glas with ice and sonicate the sample for approximately 8-10 rounds of 30 sec with 70% power and 1 min pause between each round. Take care that no air bubbles are formed during sonication and that the sample does not become warm.
- Transfer the lysed cells into SS34 centrifugation tubes and centrifuge at 20,000 rpm, 30 min, 4 °C.
- During centrifugation, wash the ÄKTA pumps and column with degassed 20% ethanol solution and degassed Millipore water and equilibrate with affinity (pump A) and elution (pump B) buffers.
- Transfer the supernatant to the superloop and connect the loop to the ÄKTA.
- Set the parameters for the ÄKTA run (pressue limit, flow, etc.) according to manufacturer recommendations. Start the run by setting the flow to 0.5 ml/min and press inject. The flow through can be collected. Switch from inject to load when the superloop is empty. Then wash with affinity buffer as long as UV reaches baseline again.
- Apply a gradient of 0- 50% elution buffer in 30 min and collect the flow through in 2 ml fractions. The protein of interest should elute during this gradient or when 100% elution buffer is applied to the column. Afterwards the ÄKTA system and column should be washed with degassed Millipore water and degassed 20% ethanol solution.
- The fractions containing the protein of interest can be analyzed by SDS PAGE. Therefore, take 20 µl samples and add 10 µl 3x loading dye, denature for 5 min at 95 °C and then load on a 12% SDS PAGE.
- Combine all fractions with proper amount of the protein of interest and concentrate them if necessary (e.g. to decrease the volume).
- Equilibrate the ÄKTA and size exclusion chromatography column in SEC buffer.
- Centrifuge the protein sample at 13,000 rpm for 10 min and inject the supernatant into the sample loop.
- Start the run with 0.35 ml/min flow and press inject. Collect the flow through and analyze peak fractions by SDS PAGE. Then combine the fractions which include the protein of interest, concentrate the solution and store at -20 °C.
- For overexpression of Aspergillus nidulans proteins in Escherichia coli (here: Rosetta DE3) inoculate 5 ml LB pre-culture, supplemented with appropriate antibiotics, for overnight incubation at 37 °C with agitation.
- Preparation of protein crude extracts
- Take 100 ml autoclaved minimal media (MM) in 300 ml buffled falsks and add supplements according to the A. nidulans strains. Inoculate with 1 x 106 spores/ml medium and incubate at 37 °C for 20 h under illumination and 120-150 rpm agitation.
- Collect the mycelium in funnels with miracloth filters. Wash the mycelium with 50-100 ml 0.96% NaCl solution and then take the miracloth filter containing the mycelium and dry it between paper towels.
- Transfer the dry mycelium (take mycel of the size of a 1 cent coin) to a 2 ml reaction tube and put into liquid nitrogen.
- Pre-cool the reaction tube holders of the Mixer Mill in liquid nitrogen by pouring a small amount into a Styrofoam box.
- Add two metal balls to each tube and put the tube into the holder.
- Close the holder and assemble them to the Mixer Mill. Set the program to 1 min and frequency to 30/sec then press start.
- Transfer the tube holder again into the box with a little bit of liquid nitrogen to prevent thawing.
- Take out the metal balls with a strong magnet. Then transfer the grinded mycelium to a new 2 ml tube and put the tubes again to liquid nitrogen.
- Put all tubes on ice and open them immediately. Add 300 µl B* buffer and vortex for ~1 min.
- Centrifuge for 30 min, 13,000 rpm, 4 °C.
- Transfer the supernatant to a fresh 1.5 ml tube and centrifuge again for 10 min, 13,000 rpm, 4 °C. In case there is still a pellet, transfer the supernatant to a fresh 1.5 ml tube.
- Measure the protein concentration with Nanodrop. To prevent protein aggregate formation, do not store the crude extracts longer than 1 week at -20 °C (to perform the deneddylation assay at the same or next day would be optimal).
- Take 100 ml autoclaved minimal media (MM) in 300 ml buffled falsks and add supplements according to the A. nidulans strains. Inoculate with 1 x 106 spores/ml medium and incubate at 37 °C for 20 h under illumination and 120-150 rpm agitation.
- Deneddylation assay
- Protein crude extracts of csn deletion and wild type strains serve as neddylated and deneddylated cullin source. Recombinant A. nidulans CSN proteins are used to complement the deletions. If necessary, thaw all protein solutions, centrifuge at 13,000 rpm for 10 min at 4 °C (do not freeze and thaw protein solutions more than 2-3 times as proteins form aggregates). Transfer the supernatant to a new tube. Re-measure the concentration using Nanodrop.
- For each reaction, mix 80 µg of the protein crude extracts with 8 µg recombinant protein, 100 mM DTT and B* buffer to a final volume of 30 µl in a 1.5 ml reaction tube (Table 1). As a control, prepare the same reactions without recombinant protein. Incubate at 37 °C for 30 min and 120 rpm agitation.
Table 1. Pipetting scheme (example)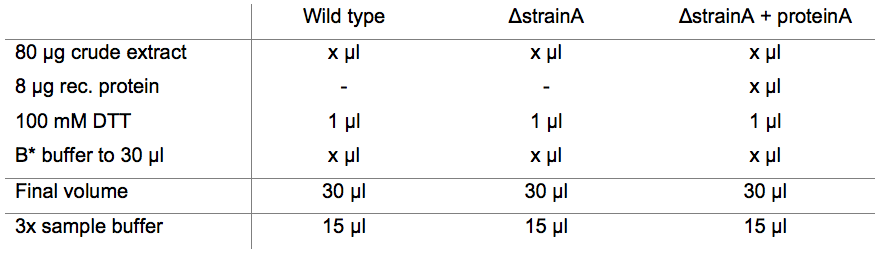
- Add 15 µl 3x SDS sample buffer to each reaction, mix, denature at 95 °C for 5 min and then cool down on ice for 1 min. Flash spin the samples for 3-5 sec in a table centrifuge.
- Prepare 12% SDS gels.
- Load 10 µl of each sample on a gel and use 3.5 µl of Page Ruler Prestained Protein Ladder as size standard. Use 1x running buffer to run the electrophoresis. Apply 120 V until the proteins reach the resolving gel and then increase the voltage to 150-180 V until the 25 kDa band of the page ruler ran out of the gel.
- Stop the migration and carefully transfer the gel to the western blotting apparatus. Assemble the western blot according to the manufacturer recommendations and as represented in Figure 1. Close the cassette with the colorless site and transfer it into the Mini trans-blot central core after it was placed into the electrophoresis chamber filled with transfer buffer. The proteins can be blotted either for at least 1 h at 100 V and appropriate cooling of the buffer or overnight at 35 V.
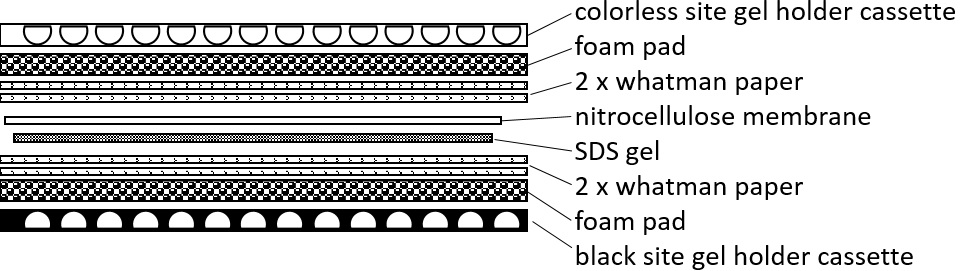
Figure 1. Western blot assembly. Schematic representation of the order of blot assembly for western hybridization. - Disassemble the blot and cover the membrane with blocking solution for 1 h at room temperature and gentle agitation.
- Discard the blocking solution and separate the membrane into two pieces at the 70 kDa page ruler band (Figure 2). Add the primary antibody α-Nedd8 or α-CulA (dilution in blocking solution 1:1,000) to the upper membrane. As loading control the primary antibody α-Tub or α-Act (dilution in blocking solution 1:1,000) can be added to the lower membrane part. Incubate for 1 h at room temperature and gentle agitation.
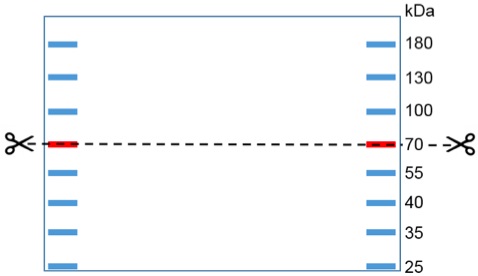
Figure 2. Western blot membrane. The dashed line indicates where the membrane needs to be cut in order to develop the loading control (Actin or Tubulin ~40 kDa) simultaneously with the CulA or Nedd8 antibody (signal ~100 kDa). - Remove the primary antibodies and wash the membranes 3 x 10 min with 1x TBST at room temperature and gentle agitation.
- Add the secondary antibody α-goat rabbit to the upper membrane and α-goat mouse (both 1:1,000 in blocking solution) to the lower membrane for 1 h at room temperature and gentle agitation.
- Remove the secondary antibodies and wash the membranes 3 x 10 min with 1x TBST buffer at room temperature and gentle agitation.
- For signal detection prepare the detection solutions freshly. Then mix detection solution A with detection solution B on the membrane for 2 min at room temperature and gentle agitation. Subsequently place the membrane into a plastic foil and transport it via an autoradiography cassette to the Fusion imaging system (important: switch on the PC and Fusion imaging system before you are ready to use, because the camera needs approximately 10 min for cooling down).
- Click on ‘start preview’, place the membrane into the apparatus, click ‘Set-up’ → ‘Sensitivity’ → ‘Super Sensitivity’ → ‘Auto Exposure’ (takes 1-4 min to detect appropriate signals). Save the image as TIF file (also automatically saved as STB file).
- Analyze signals with Fusion BIO-1D software as followed:
- Click ‘Open an Image’ and then choose the image. Click ‘Optical Density’.
- A- Lane definition: click ‘New Lane’, a box with green lines appears (Figure 3). Drag around the signals to define the area to be analyzed. click ‘Select all’ → ‘Next >>’.
- B- Background subtraction: click ‘Do rolling ball’ → ‘Next >>’.
- C- Spot separation: click ‘Add separation’ or use given separations and place the brackets around band boundaries by dragging the cursor (Figure 4). Click ‘Next >>’.
- Untick ‘Concentration’, ‘Height’, Area’ and ‘Molecular Weight’ (only ‘Volume’ has a tick) (Figure 5). Press the ‘Excel’ button. An Excel window opens with the volume data representing the pixel intensity inside a defined boundary (Figure 6). Further quantify signals considering wild type signal and loading controls.
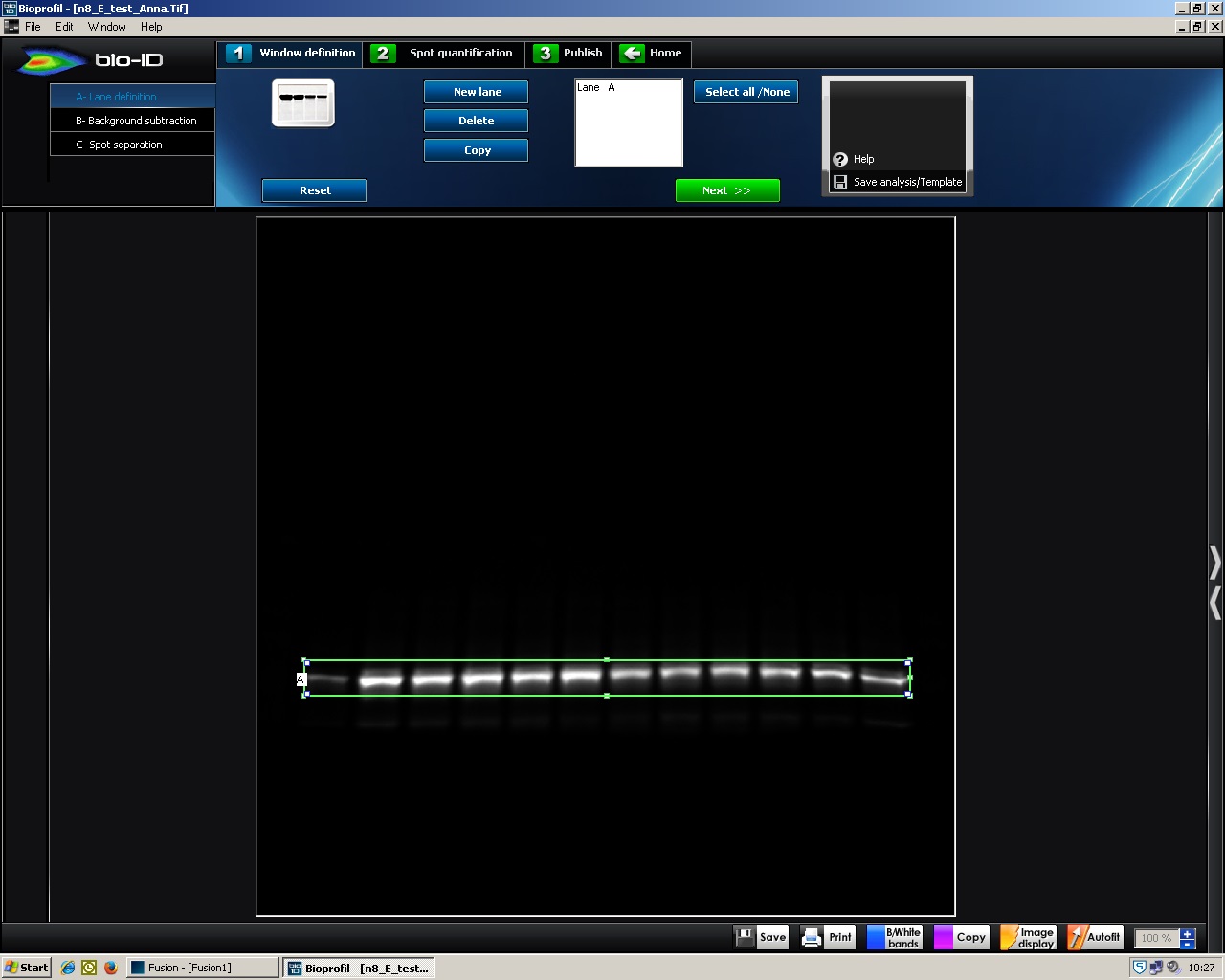
Figure 3. BIO-1D, A-Lane definition. Screenshot of BIO-1D software showing a green lined box that can be dragged to define the analysis area.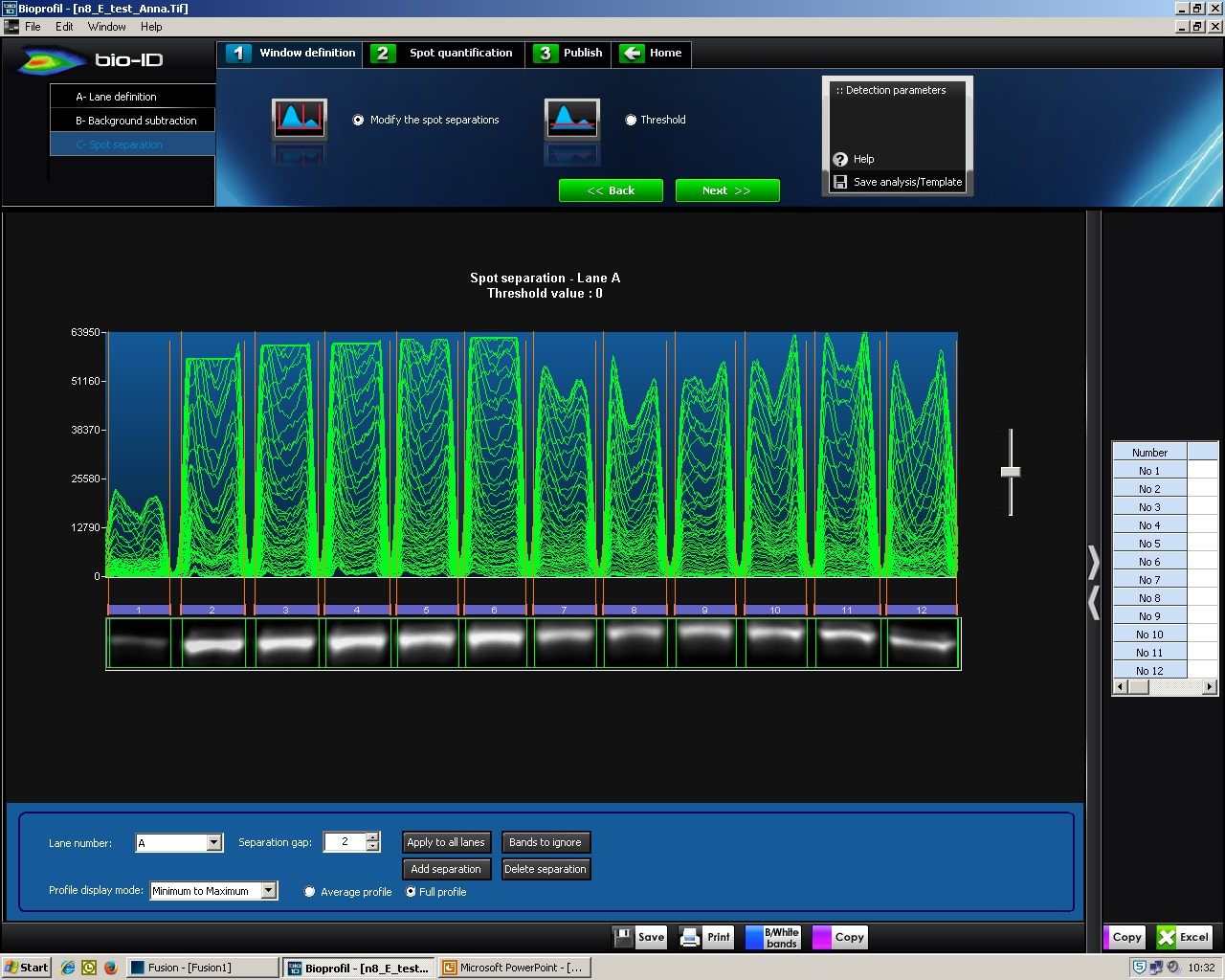
Figure 4. BIO-1D, C-Spot separation. Screenshot of BIO-1D software displaying the separation of the band boundaries by placing brackets.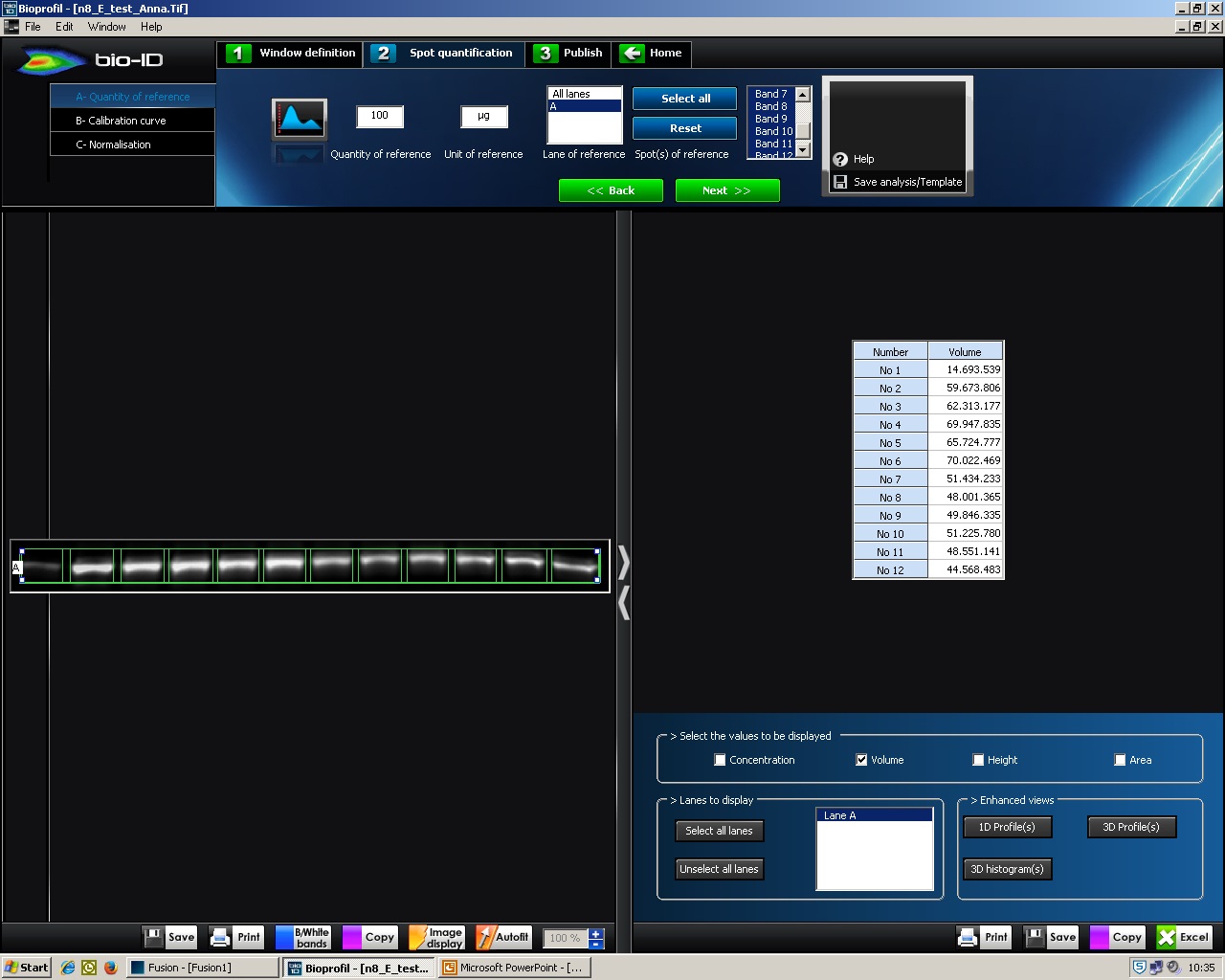
Figure 5. BIO-1D, Data export to Excel. Screenshot of BIO-1D software displaying the step where the value of the Volume has to be selected to be exported to Excel.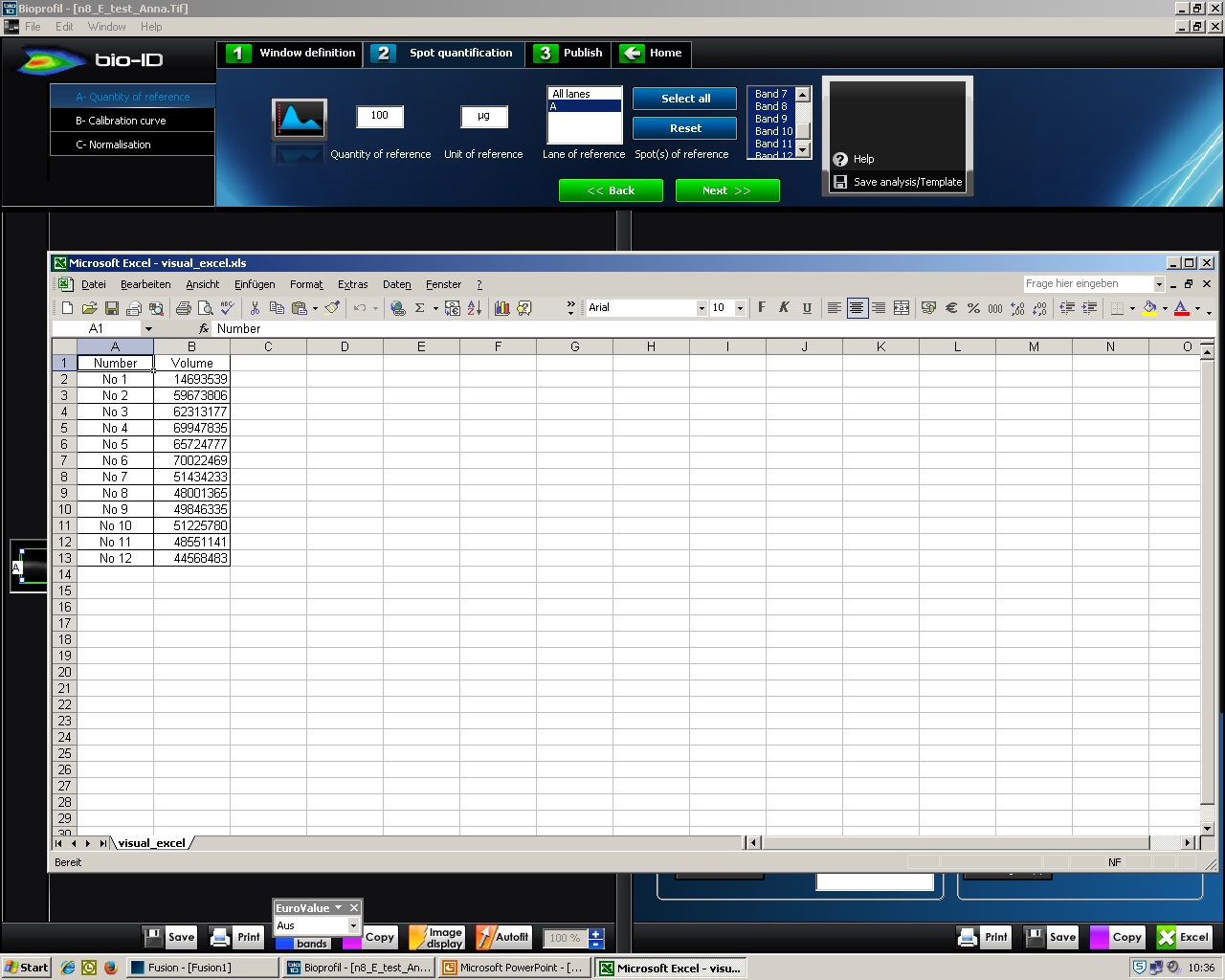
Figure 6. BIO-1D, Data in Excel. Screenshot of Excel table with values of pixel intensity inside a defined boundary. - Click ‘Open an Image’ and then choose the image. Click ‘Optical Density’.
- Protein crude extracts of csn deletion and wild type strains serve as neddylated and deneddylated cullin source. Recombinant A. nidulans CSN proteins are used to complement the deletions. If necessary, thaw all protein solutions, centrifuge at 13,000 rpm for 10 min at 4 °C (do not freeze and thaw protein solutions more than 2-3 times as proteins form aggregates). Transfer the supernatant to a new tube. Re-measure the concentration using Nanodrop.
Representative data
Figure 7 shows a typical result of an in vitro deneddylation assay similar to those shown in Beckmann et al. (2015). Usually most cullins are deneddylated in wild type. This is visible in the western hybridization and quantification of crude extracts from wild type (wt) where the Nedd8 antibody gives a weak signal at ~100 kDa, with CulA hybridization a strong signal for deneddylated CulA (CulA, **) and a slight signal for neddylated CulA (CulA-Nedd8, *) (Figure 7). In contrast, crude extracts of csn deletion strains show increased neddylated CulA amounts, which cannot be complemented by the addition of the respective recombinant CSN subunit to the deletion strain. The only exception where CulA is mostly deneddylated like in wild type is when recombinant CsnE is added to csnE deletion crude extract. Only then the CSN seems to be active.
Signals were quantified relative to the loading control and for Nedd8 relative to wild type. For CulA signals the ratio of neddylated CulA to non-neddylated CulA were calculated.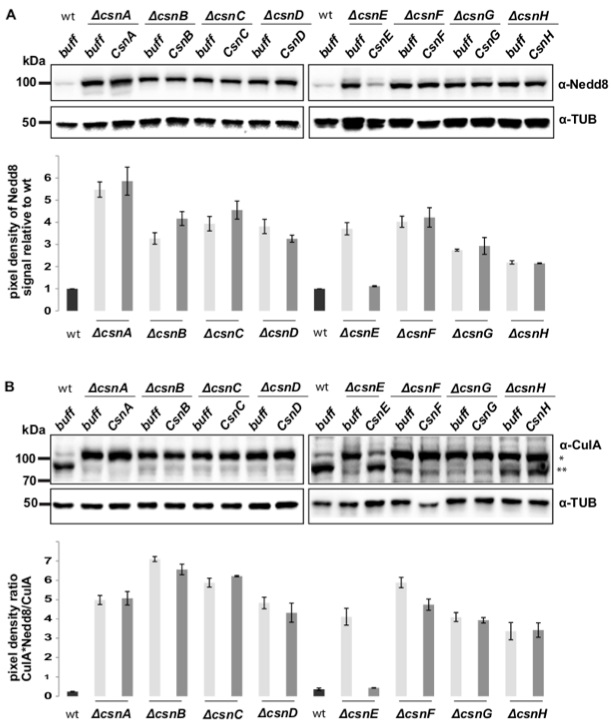
Figure 7. An example for in vitro deneddylation assay: CsnE can restore deneddylation activity of CSN. Western hybridization of A. nidulans crude extracts incubated with and without recombinant CSN proteins. A. The membrane was incubated with α-Nedd8 antibody resulting in a weak signal for wt and ΔcsnE + CsnE and strong signals for all other csn deletion strains with or without recombinant protein at ~100 kDa. α-Tub antibody was used as loading control. B. Detection with α-CulA underlined the result from (A) where the ratio of neddylated CulA (*) to deneddylated CulA (**) in ΔcsnE + recombinant CsnE was similar to the wild type and inverted in comparison to all other samples. Results were quantified using BIO1D software (Peqlab).
Notes
Users of this protocol should have experience in or collaborative groups for protein purification using the ÄKTA system.
For reliable data, at least three biological repetitions should be performed. This is also important for calculating error bars. Mean values and standard deviation were calculated using Excel with the formula below: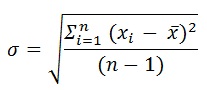
With σ as standard deviation, xi representing each value of the dataset, x̄ arithmetic mean value, n total number of data points, Σ the sum of (xi - x̄)2 of all data points.
Recipes
- Lysogeny broth (LB) medium
1% (w/v) Peptone/Tryptone
0.5% (w/v) yeast extract
1% (w/v) NaCl
Autoclave and supplement with antiobiotics (1:1,000) before usage. - Lysis buffer
500 mM NaCl
50 mM Tris (pH 8.0)
Add just before use: 1 mM PMSF - Affinity buffer
500 mM NaCl
50 mM Tris (pH 8.0)
5% (w/v) Imidazole in case of HisTrap columns - Elution buffer for HisTrap
500 NaCl
50 mM Tris (pH 8.0)
500 mM Imidazole - Elution buffer for GSTrap
500 NaCl
50 mM Tris (pH 8.0)
20 mM Glutathione - Size exclusion buffer (SEC)
120 mM NaCl
20 mM Tris pH 8.0
1 mM DTT
1 mM EDTA
5% (v/v) Glycerol - Minimal medium
1% (w/v) Glucose
1x AspA
2 mM MgSO4
1x trace elements - 50x AspA
3.5 M NaNO3
350 mM KCl
560 mM KH2PO4
Adjust to pH 5.5 with KOH (~30 mL 5 M KOH needed!), autoclave - Trace elements
18 µM FeSO4
174 µM EDTA
76 µM ZnSO4
178 µM H3BO3
25 µM MnCl2
7.1 µM CoCl2
6.4 µM CuSO4
6.2 µM Na2MoO4
Sterile filter - B* buffer
300 mM NaCl
10 mM Tris-HCl (pH 7.5)
0.5 mM EDTA
0.2% (v/v) NP-40
10% (v/v) Glycerol
Add just before use:
1 mM PMSF
2 mM DTT
5 mM E64
100 µl/10 ml Complete Protease Inhibitor Cocktail - 3x SDS sample buffer
250 mM Tris-HCl (pH 6.8)
15% 2-mercaptoethanol
30% Glycerol
7% SDS
0.3% Bromophenol blue - 12% SDS gels
for 2 gels mix the following solutions in a 50 ml reaction tube:
Resolving gel:
5 ml 2x resolving buffer
4 ml 30% acrylamide
1 ml H2O
150 µl 10% APS
15 µl TEMED- Invert the solution and then fill between the glas plates.
- Add ~1 ml 100% isopropanol on top of the gel to prevent air bubbles.
- Let the gels dry ~10-20 min.
- Then remove the isopropanol carefully.
- Use whatman paper to remove remaining drops.
- Then mix all components for the stacking gel in a 50 ml reaction tube.
2 ml 2x stacking buffer
0.5 ml 30% acrylamide
1.5 ml H2O
60 µl 10% APS
7.5 µl TEMED- Invert the tube and then fill the stacking solution onto the resolving gel up to the top.
- Then place the comps carefully into the stacking gel.
- Let the gels dry for another 10-20 min.
- 2x resolving buffer
0.75 M Tris-HCl (pH 8.8)
0.1% (w/v) SDS - 2x stacking buffer
0.25 M Tris-HCl (pH 6.8)
0.1% (w/v) SDS - 10x SDS running buffer
250 mM Tris-base
2.5 M Glycine
1% (w/v) SDS
Dilute the buffer to 1x working solution with ddH2O - 10 x transfer buffer
250 mM Tris-base
1.92 M glycine
0.2% (w/v) SDS
Dilute the buffer to 1x working solution with ddH2O - 10x TBST (Tris-Buffered Saline Tween 20)
100 mM Tris-HCl (pH 8.0)
1.5 M NaCl
0.5% (v/v) Tween-20
Dilute the buffer to 1 x working solution with ddH2O - Blocking solution
5 % milk powder dissolved in 1x TBST buffer - Detection solution A (prepare fresh just before use)
9 ml ddH2O
1 ml Tris-HCl (pH 8.5)
100 µl 250 mM Luminol (dissolve stock solution in DMSO, protect from light, store at -20 °C)
44 µl 400 mM p-coumaric acid (dissolve stock solution in DMSO, protect from light, store at -20 °C) - Detection solution B (prepare fresh just before use)
9 ml ddH2O
1 ml 1 M Tris-HCl (pH 8.5)
6.15 µl 30% H2O2 - Complete Protease Inhibitor Cocktail
Dissolve 1 ½ tablets in 1 ml B*buffer
Acknowledgments
This protocol was adapted from a previously published study (Beckmann et al., 2015) and precursor experiments (Christmann et al., 2013; Harting et al., 2013; von Zeska Kress et al., 2012). We thank Dr. Elena Beckmann for her support and advice and Gabriele Heinrich for excellent technical assistance. We also thank Sabine Reen and Josua Schinke for careful proofreading the manuscript. This research has been supported by the Deutsche Forschungsgemeinschaft (DFG) within the SFB860.
References
- Beckmann, E. A., Köhler, A. M., Meister, C., Christmann, M., Draht, O. W., Rakebrandt, N., Valerius, O. and Braus, G. H. (2015). Integration of the catalytic subunit activates deneddylase activity in vivo as final step in fungal COP9 signalosome assembly. Mol Microbiol 97(1): 110-124.
- Christmann, M., Schmaler, T., Gordon, C., Huang, X., Bayram, Ö., Schinke, J., Stumpf, S., Dubiel, W. and Braus, G. H. (2013). Control of multicellular development by the physically interacting deneddylases DEN1/DenA and COP9 signalosome. PLoS Genet 9(2): e1003275.
- Harting, R., Bayram, Ö., Laubinger, K., Valerius, O. and Braus, G. H. (2013). Interplay of the fungal sumoylation network for control of multicellular development. Mol Microbiol 90(5): 1125-1145.
- von Zeska Kress, M. R., Harting, R., Bayram, Ö., Christmann, M., Irmer, H., Valerius, O., Schinke, J., Goldman, G. H. and Braus, G. H. (2012). The COP9 signalosome counteracts the accumulation of cullin SCF ubiquitin E3 RING ligases during fungal development. Mol Microbiol 83(6): 1162-1177.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Köhler, A. M., Meister, C. and Braus, G. H. (2016). In vitro Deneddylation Assay. Bio-protocol 6(6): e1756. DOI: 10.21769/BioProtoc.1756.
Category
Microbiology > Microbial biochemistry > Protein > Activity
Microbiology > Microbial biochemistry > Protein > Modification
Biochemistry > Protein > Modification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










