- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Isolation, Culture, and Maintenance of Mouse Intestinal Stem Cells
Published: Vol 6, Iss 4, Feb 20, 2016 DOI: 10.21769/BioProtoc.1733 Views: 30273
Reviewed by: Xuecai GeAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Preparing Chamber Slides With Pressed Collagen for Live Imaging Monolayers of Primary Human Intestinal Stem Cells
Joseph Burclaff and Scott T. Magness
Nov 20, 2024 2030 Views

Cochlear Organ Dissection, Immunostaining, and Confocal Imaging in Mice
Chenyu Chen [...] Dongdong Ren
Jan 20, 2025 3773 Views

Isolation and Imaging of Microvessels From Brain Tissue
Josephine K. Buff [...] Sophia M. Shi
Aug 5, 2025 2637 Views
Abstract
In this protocol we describe our modifications to a method to isolate, culture and maintain mouse intestinal stem cells as crypt-villus forming organoids. These cells, isolated either from the small or large intestine, maintain self-renewal and multilineage differentiation potential over time. This provides investigators a tool to culture wild type or transformed intestinal epithelium, and a robust assay for stem cell tissue homeostasis in vitro.
Keywords: OrganoidMaterials and Reagents
- Cover glass (Corning, catalog number: 2998075X25 )
- 48-Well Tissue Culture Plate (Corning, Falcon®, catalog number: 351178 )
- 50 ml and 15 ml Conical centrifuge tubes (Corning, Falcon®, catalog number: 352098 and 352097 )
- 10 ml Syringe (BD, catalog number: 309604 )
- 21G Needle (BD, catalog number: 305165 )
- Pipette Tips
- 70 μM Cell Strainer (Corning, Falcon®, catalog number: 352350 )
- 100 μM Cell Strainer (Corning, Falcon®, catalog number: 352360 )
- Mice to be harvested for this protocol must be approved for use by the Institutional Animal Care and Use Committee (IACUC) at the institution, which sponsors the laboratory research
- Phosphate Buffered Saline (PBS) (Invitrogen, catalog number: 10010023 )
Note: Currently, it is “ Thermo Fisher Scientific, GibcoTM, catalog number: 10010023”. - Growth Factor Reduced Matrigel (BD, catalog number: 356230 )
Note: Currently, it is “Corning, Matrigel®, catalog number: 356230”. - Fetal Bovine Serum (Thermo Fisher Scientific, GibcoTM, catalog number: 16000-044 )
- Collagenase Type IV (Worthington, catalog number: LS004188 )
- Bovine Serum Albumin (Sigma-Aldrich, catalog number: A2058 )
- 1% BSA-PBS (sterile)
- Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: E9884 )
- 5 mM EDTA-PBS
- Recombinant DNase I, RNase Free (which is provided at 10 U/μl) (Sigma-Aldrich, catalog number: 4716728001 )
- Optional: 10 μM Rho Kinase Inhibitor Y-27632, provided as a 5 mM Solution (Merck Millipore Corporation, catalog number: 68801 )
- Advanced DMEM F/12 (Thermo Fisher Scientific, GibcoTM, catalog number: 12634-010 )
- Streptomycin (Gibco, catalog number: 15140-22 )
- N-Acetylcysteine (Sigma-Aldrich, catalog number: A9165 )
- HEPES (Sigma-Aldrich, catalog number: H3375 )
- EGF 50 ng/ml (Invitrogen, catalog number: PMG8043 )
Note: Currently, it is “ Thermo Fisher Scientific, GibcoTM, catalog number: PMG8043”. - Recombinant Murine EGF 50 ng/ml (Invitrogen, catalog number: PMG8043)
- Recombinant Murine Noggin 50 ng/ml (Peprotech, catalog number: 250-38 )
- Recombinant Human R-Spondin 1,500 ng/ml (R&D Systems, catalog number: 3474-RS-050 )
- Recombinant Murine Wnt-3A 100 ng/ml (Merck Millipore Corporation, catalog number: GF-160 )
- 10 mM Nicotinamide (Sigma-Aldrich, catalog number: N3376 )
- Intestinal basal medium (see Recipes)
- Small intestinal organoid growth media (see Recipes)
- Large intestinal organoid growth media (see Recipes)
Equipment
- Pipettes, Pipetaid and Micro-pipettes
- Dissection Forceps and Scissors (Fine Science Tools, catalog number: 11150-10 and 14058-09 )
- Centrifuge 5810R (Eppendorf, model: 5810R )
- Brightfield inverted Microscope
- Tabletop Roller in 4 °C Room (Bibby Scientific Limited, Stuart Equipment, model: SRT9D )
- 37 °C, 5% CO2 cell culture incubator
- Biosafety cabinet (Tissue culture hood)
- For mouse sacrifice by carbon dioxide asphyxiation, a carbon dioxide source, regulated dispenser, and euthanasia chamber, must be used in accordance with approved animal use protocols at the laboratory’s sponsoring institution.
Procedure

Figure 1. An overview of the key steps for the isolation of intestinal crypt containing stem cells from mouse colon or small intestine
Part I. Isolation, culture, maintenance, passage and preservation of small intestinal organoids
- Procedure to isolate and culture small intestinal organoids (expected time to completion: 2-3 h)
- Mice are euthanized by carbon dioxide asphyxiation as recommended by the 2000 Report of the AVMA Panel on Euthanasia (Association, 2001).
- Dissect and remove 15 cm of the proximal small intestine. Be careful to avoid touching any of the mouse skin or hair to your tools to prevent microbial contamination. After dissected, flush the lumen of the intestine with ice-cold PBS using a 10 ml Syringe and 21G needle.
- Use scissors to open the intestine longitudinally and place in a 50 ml conical tube containing 25 ml ice-cold PBS. Invert 10-15 times, remove the PBS and replace with 25 ml of ice-cold PBS. Repeat this process until the supernatant no longer contains any visible debris.
- Cut the intestine into 5 millimeter pieces and place into 10 ml ice-cold 5 mM EDTA-PBS. Vigorously triturate the fragments by pipetting up and down into a 10 ml pipette 15 times. Let the fragments settle by gravity for 30 sec.
- Aspirate the supernatant, being careful to avoid the intestinal fragments, and replace with 10 ml, 5 mM EDTA-PBS and place at 4 °C on a benchtop roller for 10 min.
- Repeat step A5 by aspirating the supernatant, replacing with 10 ml of 5 mM EDTA-PBS, but now place at 4 °C on a benchtop roller for 30 min.
- Aspirate the supernatant, gently add 10 ml of cold PBS to wash the crypts and aspirate. Then, add 10 ml of cold PBS and then vigorously triturate with a 10 ml pipette 10 times.
- Collect this 10 ml supernatant fraction in a separate tube. Add 10 ml of cold PBS to the crypts, vigorously triturate 10 times, and then collect this fraction in a separate tube. Repeat step A7 once more to collect a total of three fractions.
- Examine 10 μl of each fraction under a microscope, looking for the presence of intact intestinal crypts and lack of larger, fragmented villi. An example of intact intestinal crypts isolated from a mouse that harbors a GFP knock-in allele at the Lgr5 locus (Barker et al., 2007) is shown in Figure 2.
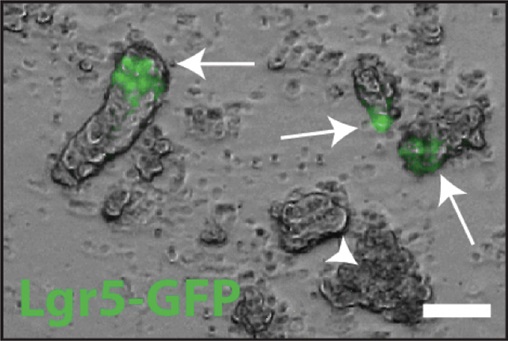
Figure 2. A brightfield/epifluorescent image overlay of freshly isolated small intestinal crypts that harbor the Lgr5-GFP knock in allele (Barker et al., 2007). Intact crypts are marked by white arrows, and contain Lgr5+ stem cells marked by the presence of GFP (green). Debris (non-viable cells) are marked by the white arrowhead. Scale bar is 50 μm. - Using the 10 ml fraction that contains the most crypts, mix with 10 ml Basal Media + 15 U/ml DNase I (add 30 μl to 10 ml Basal Media) and filter through a 100 μm filter into a BSA (1%) coated 50 ml conical tube; to coat the tube, add 1 ml of 1% BSA, shake vigorously and discard excess. Coating the tube with BSA prevents adsorption of the crypts to the sides of the tube, therefore increasing yield.
- Filter the solution again, this time through a 70 μm filter into a BSA (1%) coated tube and then spin the filtrate at 300 x g in a tabletop centrifuge for 5 min.
- Aspirate the supernatant. Depending on multiple factors (animal diet, time of day of harvest, how vigorous each washing step was performed) there can be a layer of mucous that accumulates in the supernatant, sometimes near the pellet of cells. Aspirating this will increase the purity of your yield, however it can also pull the cell pellet with it during vacuum suction. Be careful to avoid losing your pellet at this step, it is possible to use a 10 ml pipette to gently aspirate the supernatant at this step.
- Resuspend the cell pellet with 5 ml basal media containing 5% FBS and centrifuge at 100 x g for 5 min.
- Resuspend the cell pellet in 200 μl of basal media and count crypts from a 10 μl aliquot. You may have to first dilute the aliquot that you count in order to get an accurate number. Keep the volume of media that you resuspend the crypts in as low as possible because you will aim to mix 100-1,000 crypts at a 1:10 mixture with Matrigel. In a typical isolation, we recover 250 crypts/μl (in basal media) and mix this 1:10 with Matrigel.
- Plate 40 μl of the Matrigel:crypt mixture as a bubble on one well of a 48 well plate, being careful to avoid letting the Matrigel touch the edges of the well (for a schematic see Figure 3). Although organoids will still grow in Matrigel that has ‘collapsed’ to the sides of the wells, it is easier to change media if the Matrigel remains as a single bubble in the center of the well. Incubate the plate in a 37 °C incubator to allow the Matrigel to polymerize for 10-15 min.
- Add 250 μl of Small Intestinal Organoid Growth Media to each well. Over the course of 7-10 d, viable intestinal stem cells should form organoids with a crypt-villus structure (for example, see Figure 4).
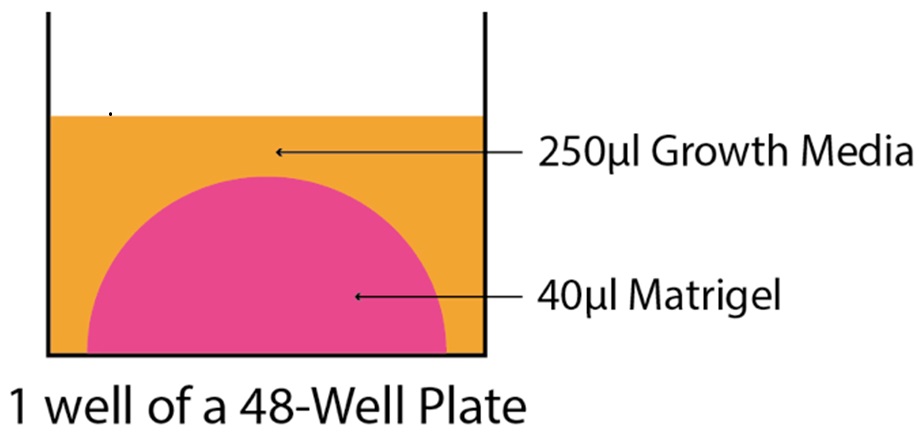
Figure 3. A schematic illustrating a properly plated 40 μl ‘Matrigel bubble’ at the bottom of one well of a 48-well plate. 250 μl of growth media is added on top of the bubble after polymerization. Ensuring that the Matrigel does not contact the well edges allows for a pipette to be placed down the side of the well to remove media during media changes.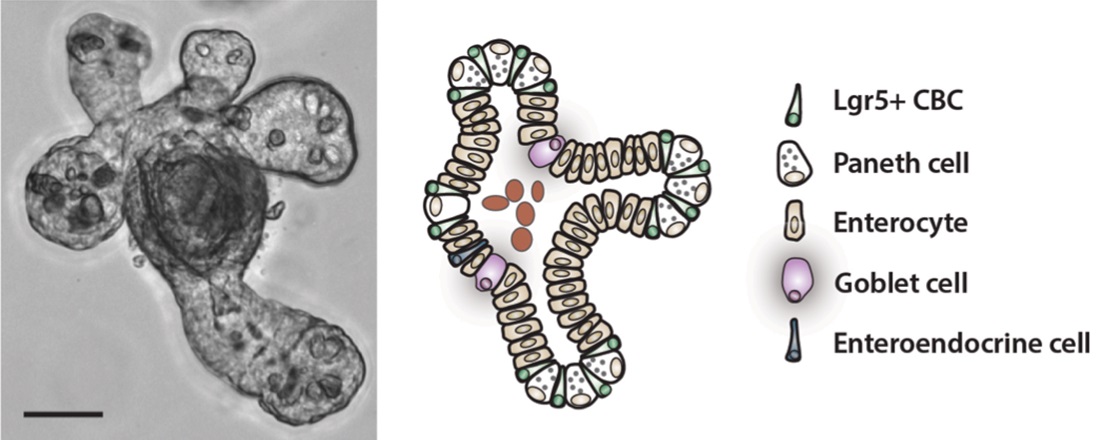
Figure 4. A high-resolution bright field microscopic picture of a small intestinal organoid, accompanied by a schematic depiction of each of the individual cell types observed in the structure. Scale bar is 50 μm.
- Mice are euthanized by carbon dioxide asphyxiation as recommended by the 2000 Report of the AVMA Panel on Euthanasia (Association, 2001).
- Procedure to maintain, passage, and cryogenically preserve small intestinal organoids
- After isolation, small intestinal growth media is changed every two days.
- After 5-7 days of growth, or when the organoids show dark, necrotic cores, they are passaged. To do this, the growth media is removed and 500 μl of cold PBS is added to the well and Matrigel is broken up by pipette, being careful to avoid bubble formation in the resuspension mixture.
- Transfer the resuspension to a 15 ml conical tube. Using a p200 pipette, pipette up and down 50-100 times to mechanically disassociate the organoids into smaller fragments. When finished, another 7 ml of cold PBS is added to the mixture and pipetted up and down 20 times.
- Centrifuge the cells at 200 x g for 5 min in a tabletop centrifuge.
- Carefully aspirate the supernatant, being careful not to remove the pellet. The pellet may not be visible to the naked eye; this is especially true if only passaging 1 well. Resuspend the pellet in Matrigel, and replate 40 μl per well into a 48 well plate. For non-transformed organoid cultures, we typically split at a 1:4 ratio.
- If preparing cells for storage in liquid nitrogen, resuspend the pellet in Basal Media containing 10% FBS and 10% DMSO. Allow to freeze slowly in a -80 °C freezer and then move to liquid nitrogen for indefinite storage.
- After isolation, small intestinal growth media is changed every two days.
Part II. The isolation, culture, maintenance, passage and cryogenic preservation of large intestinal organoids
- Procedure to isolate and culture large intestinal organoids (Video 1)Video 1. Procedure to isolate and culture large intestinal organoids
- Mice are euthanized as recommended by the 2000 Report of the AVMA Panel on Euthanasia (Association, 2001).
- Dissect and remove 5-7 cm of the proximal large intestine. Be careful to avoid touching any of the mouse skin or hair to your tools to prevent microbial contamination. After dissected, flush the lumen of the intestine with cold PBS.
- Using scissors, open the colon longitudinally. Using a glass slide, gently scrape the lumen of the intestine to remove fecal matter and mucous. Place in a 50 ml conical tube with 25 ml ice-cold PBS. Invert 10-15 times, remove the PBS and replace with 25 ml of ice-cold PBS. Repeat this process until the supernatant no longer contains any visible debris.
- Cut the intestine into 5 millimeter pieces and place into 10 ml cold 5mM EDTA-PBS. Vigorously triturate the fragments by pipetting up and down into a 10 ml pipette 15 times. Let the fragments settle by gravity for 30 sec.
- Aspirate the supernatant, being careful to avoid the intestinal fragments, and replace with 10 ml, 5 mM EDTA-PBS and place at 4 °C on a benchtop roller for 15 min.
- Remove the EDTA from the tube by aspiration. Wash 2 x with PBS. Then add 3 ml of Basal Medium containing 500 U/ml Collagenase Type IV. Pipette up and down 5 times using a 5 ml pipette. Place in a 37 °C water bath for 30 min.
- Add 10 ml of ice-cold PBS and then vigorously triturate with a 10 ml pipette 10 times.
- Collect this 10 ml supernatant fraction in a separate tube. Add 10ml of cold PBS to the crypts, vigorously triturate 10 times, and then collect this fraction in a separate tube. Repeat step B7 once more to collect a total of three fractions.
- Pipette 10 μl of each fraction on to a glass slide and examine underneath a microscope, looking for the presence of intact colonic crypts.
- To the 10 ml fraction that contains the most crypts, add 10 ml Basal Media containing 15 U/ml DNase I (add 30 μl to 10 ml Basal Media) and then filter through a 100 μm filter into a BSA (1%) coated 50 ml Falcon tube. To coat a tube, pipette 1 ml of 1% BSA into a tube and shake vigorously. Coating the tube with BSA prevents adsorption of the crypts to the sides of the tube, therefore increasing yield.
- Filter the solution again, this time through a 70 μm filter into a BSA (1%) coated tube and then spin the filtrate at 300 x g in a tabletop centrifuge for 5 min.
- Aspirate the supernatant. Depending on multiple factors (animal diet, time of day of harvest, how vigorous each washing step was performed) there can be a layer of mucous that accumulates in the supernatant, sometimes near the pellet of cells. Aspirating this will increase the purity of your yield, however it can also pull the cell pellet with it during vacuum suction. Be careful to avoid losing your pellet at this step, it’s possible to try using a 10 ml pipette with a pipette aid to aspirate the supernatant here, or by using a p1000 pipette.
- Resuspend the cell pellet with 5 ml Basal Media containing 5% FBS and centrifuged at 100 x g for 5 min.
- Resuspend the cell pellet in 200 μl of Basal Media and count crypts from a 10 μl aliquot (for an example, see Figure 5). You may have to first dilute the aliquot that you count in order to get an accurate number. You want to keep the volume of media that you resuspend the crypts in as low as possible because you will aim to mix 100-1,000 crypts at a 1:10 mixture with Matrigel. In a typical isolation, we will resuspend 50 colon crypts/μl of basal media and mix this 1:10 with Matrigel.
- 40 μl of the resuspended mixture is plated as a bubble on the bottom of a well in a 48 well plate, being careful to avoid letting the Matrigel touch the edges of the well (see Figure 3). The plate is placed in a 37 °C incubator to allow the Matrigel to polymerize for 10-15 min.
- Add 250 μl of Large Intestinal Organoid Growth Media to each well.
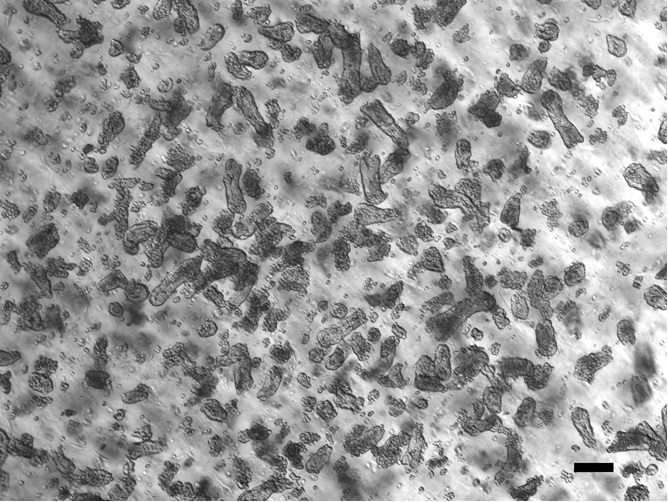
Figure 5. A bright field image of a 10 μl aliquot from the 200 μl resuspension of colon crypts in step A14 of Part II. Scale bar is 50 μm.
- Mice are euthanized as recommended by the 2000 Report of the AVMA Panel on Euthanasia (Association, 2001).
- Maintenance, passage, and cryogenic preservation of colon organoids
This part is performed exactly as described for the colon organoids as it is for the small intestinal organoids.
Notes
- We typically test 2 or 3 lots of Matrigel and reserve a large quantity of the best performing lot. To test a lot, we plate freshly isolated stem cells into Matrigel and observe their growth after two weeks, including one passage.
- R-spondin1 and Wnt3a producing cell lines can be obtained from investigators who have made them previously (Sato et al., 2011) in order to condition media with these respective growth factors, greatly reducing costs.
- Noggin is typically added to intestinal stem cell culture growth media at a final concentration of 100 ng/ml, but we have found that 50 ng/ml is enough to support normal stem cell growth, differentiation and self-renewal.
- All of the growth factors that are bought commercially can be dissolved in 0.1% BSA-PBS and stored as concentrated stock solutions at -20 °C.
- When preparing the Collagenase Type IV (for colon organoid isolation), make a fresh 10x stock before starting. The powder is static, so weigh it on a piece of aluminum foil and place in a 15 ml conical tube, add basal media to bring to the correct concentration, and make sure everything goes into solution by vortexing or incubating at 37 °C. Then filter sterilize through a 0.45 μm filter.
- 10 μM Rho Kinase Inhibitor Y-27632 can be added to the growth media to increase the yield of organoid growth, although we routinely isolate and grow organoids without this inhibitor.
- Colon organoids should begin to grow as large hollow spheres 4-6 days after isolation, note that this is longer than typically observed for small intestinal stem cell growth.
Recipes
- Intestinal basal medium
Supplement a 500 ml bottle of advanced DMEM F/12 with
2 mM L-Glutamine
100 units/ml Penicillin, 1,000 μg/ml Streptomycin
1 mM N-acetylcysteine
10 mM HEPES - Small intestinal organoid growth media
Supplement the basal medium with
Recombinant Murine EGF 50 ng/ml
Recombinant Murine Noggin 50 ng/ml
Recombinant Human R-Spondin-1 500 ng/ml - Large intestinal organoid growth media
Supplement the basal medium with
Recombinant murine EGF 50 ng/ml
Recombinant murine noggin 50 ng/ml
Recombinant human R-Spondin-1 500 ng/ml
Recombinant murine Wnt-3A 100 ng/ml (Millipore GF-160)
10 mM nicotinamide
Acknowledgments
This protocol was adapted from (Sato et al., 2009) with help from Caroline Lindemans. This work was supported by a program project grant from the NIH/NCI (CA-013106). L. E. D. was supported by a National Health and Medical Research Council (NHMRC) Overseas Biomedical Fellowship and a K22 Career Development Award from the NCI/NIH (CA 181280-01). K. P. O. was supported by a Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the NIH under award number T32GM07739 to the Weill Cornell/Rockefeller/Sloan-Kettering Tri-Institutional MD-PhD Program. S. W. L. is the Geoffrey Beene Chair of Cancer Biology and an investigator of the Howard Hughes Medical Institute.
References
- Association, A. P. o. E. A. V. M. (2001). 2000 report of the AVMA panel on euthanasia. J Am Vet Med Assoc 218(5): 669-696.
- Barker, N., van Es, J. H., Kuipers, J., Kujala, P., van den Born, M., Cozijnsen, M., Haegebarth, A., Korving, J., Begthel, H., Peters, P. J. and Clevers, H. (2007). Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449(7165): 1003-1007.
- Sato, T., Stange, D. E., Ferrante, M., Vries, R. G., Van Es, J. H., Van den Brink, S., Van Houdt, W. J., Pronk, A., Van Gorp, J., Siersema, P. D. and Clevers, H. (2011). Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology 141(5): 1762-1772.
- Sato, T., Vries, R. G., Snippert, H. J., van de Wetering, M., Barker, N., Stange, D. E., van Es, J. H., Abo, A., Kujala, P., Peters, P. J. and Clevers, H. (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459(7244): 262-265.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
O’Rourke, K. P., Ackerman, S., Dow, L. E. and Lowe, S. W. (2016). Isolation, Culture, and Maintenance of Mouse Intestinal Stem Cells. Bio-protocol 6(4): e1733. DOI: 10.21769/BioProtoc.1733.
Category
Stem Cell > Adult stem cell > Intestinal stem cell
Cell Biology > Tissue analysis > Tissue isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link