- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Design and Functional Analysis of Fluorescent Nitrate and Peptide Transporter Activity Sensors in Yeast Cultures
Published: Vol 6, Iss 3, Feb 5, 2016 DOI: 10.21769/BioProtoc.1728 Views: 11539
Reviewed by: Fanglian HeChong He

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Fabrication of Microfluidic Devices for Continuously Monitoring Yeast Aging
Richard O’Laughlin [...] Nan Hao
Aug 5, 2023 1449 Views

Purification of Human Cytoplasmic Actins From Saccharomyces cerevisiae
Brian K. Haarer [...] Jessica L. Henty-Ridilla
Dec 5, 2023 1753 Views

Imaging the Entire Sexual Life Cycle of the Budding Yeast Saccharomyces cerevisiae Using a Microfluidic Platform
Taylor Kennedy [...] Orlando Argüello-Miranda
Dec 5, 2025 1477 Views
Abstract
This protocol describes the methods used to engineer and deploy genetically encoded fluorescence activity reporters for nitrate and peptide transporter activity in yeast cells. Fusion of the dual-affinity nitrate transceptor CHL1/AtNRT1.1/AtNPF6.3 or four different peptide transporters (AtPTR1, 2, 4, and 5) from Arabidopsis to a pair of fluorescent proteins with different spectral properties, enabled us to engineer the NiTracs (nitrate transporter activity tracking sensors) and the PepTracs (peptide transporter activity tracking sensors), ratiometric fluorescence activity sensors that monitor the activity of the plasma membrane nitrate transceptor or the peptide transporters in vivo (Ho et al., 2014). The NiTrac1 sensor responds specifically and reversibly to the addition of nitrate, while the PepTracs respond to addition of dipeptides, either by a reduction in donor and acceptor emission, while acceptor-excited emission remains unaltered, or by a change in ratio of the fluorophore emission. All sensors are suitable for ratiometric imaging. The similarity of the biphasic kinetics of the NiTrac1 sensor response [from µM to mM (Liu and Tsay, 2003)] and the nitrate transport kinetics of the native nitrate transceptor, intimates that NiTrac1 provides information on conformational rearrangements during the transport cycle, thereby reporting transporter activity over a wide range of external nitrate concentrations. Several variants of NiTrac have been engineered, which differ with respect to their affinity for nitrate (NiTrac1: CHL1; NiTracT101A: CHL1T101A). NiTrac also recognizes chlorate. Here we describe a simple method for the design, implementation, and detection of nitrate transceptor activity in yeast cells using a spectrofluorimeter.
Keywords: BiosensorMaterials and Reagents
- Monochromator-based spectrofluorimeter for 96 well plates [for instance: Safire or Infinite® M1000 (Tecan Trading AG)]
- 96 well microplates (flat bottom clear or black) (Greiner Bio-One GmbH, catalog numbers: 650101 and 650209 )
Note: Black plates are more expensive, but have lower background. - Multichannel (12) pipette (for 100 µl) (e.g., Sartorius AG, catalog number: 725240 )
- 50 ml sterile plastic tubes (for instance: Falcon®, or any other brand)
- Yeast Strain: protease-deficient yeast strain BJ5465 [MATa, ura3–52, trp1, leu2Δ1, his3Δ200, pep4::HIS3, prb1Δ1.6R, can1, GAL+] (ATCC, catalog number: 208289TM ), which was obtained from the Yeast Genetic Stock Center (University of California, Berkeley, CA)
- The nitrate transceptor CHL1/NRT1.1/NPF6.4 (Ho et al., 2009; Leran et al., 2013) or various PTR peptide transporters (Komarova et al., 2012; Tsay et al., 2007; Leran et al., 2013)
Note: they were used as sensory domains for creating the nitrate (NiTracs) and peptide (PepTracs) sensor constructs. For this full length ORFs of CHL1, CHL1T101A, PTR1, PTR2, PTR4, and PTR5 from Arabidopsis (The Arabidopsis Information Resource) were cloned in the pTOPO Gateway Entry vector. - Sensors: CHL1/PTRs
Note: it was sandwiched between a yellow acceptor [Aphrodite t9: Aphrodite is a codon diversified Venus gene; t9 corresponds to a deletion of the C-terminus of 9 amino acids (Deuschle et al., 2006) and cyan donor fluorophore (mCerulean) (Rizzo et al., 2006)]. This was achieved by inserting the sensory domain in the Gateway yeast expression vector pDRFlip30.- pDRFlip30-NiTrac1 (original dual-affinity sensor; Km ~ 75 μM and Km ~ 3.8 Mm) (Ho et al., 2014).
- pDRFlip30-NiTrac1T101A (Variant with low-affinity; Km ~ 3.5 mM) (Ho et al., 2014).
- pDRFlip30-PepTrac1 (PepTrac1 based on AtPTR1) (Ho et al., 2014)
- pDRFlip30-PepTrac2 (PepTrac2 based on AtPTR2) (Ho et al., 2014)
- pDRFlip30-PepTrac4 (PepTrac4 based on AtPTR4) (Ho et al., 2014)
- pDRFlip30-PepTrac5 (PepTrac5 based on AtPTR5) (Ho et al., 2014)
- pDRFlip30-NiTrac1 (original dual-affinity sensor; Km ~ 75 μM and Km ~ 3.8 Mm) (Ho et al., 2014).
- Gly-Gly (Sigma-Aldrich, catalog number: G1002 ) or other di-/tri-peptides (Sigma-Aldrich)
- Potassium nitrate (Sigma-Aldrich, catalog number: P8394 )
- YNB, yeast nitrogen base w/o amino acids w/o ammonium sulfate (BD, Difco, catalog number: 233520 )
- D-(+)-Glucose monohydrate (Fluka Analytical, catalog number: 49159 )
- Agar (Sigma-Aldrich, catalog number: A1296 )
- Agarose (Sigma-Aldrich, catalog number: 05066 )
- MES hydrate (Sigma-Aldrich, catalog number: M2933 )
- Sodium hydroxide (NaOH) (Sigma-Aldrich, catalog number: S5881 )
- MilliQ or distilled water
- 1, 4-Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: DTT-RO )
- Carrier DNA [UltraPureTM Salmon Sperm DNA Solution (Thermo Fisher Scientific, InvitrogenTM, catalog number: 115632-011 )]
- Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: E6758 )
- KNO3
- KCl
- Stock
- 45% PEG4000
- Lithium acetate
- Tris-Cl (pH 7.5)
- 40x glucose solution
- -ura DropOut medium
(Takara Bio Company, Clontech, catalog number: 630416 ) (see Recipes)
- Wash buffer (see Recipes)
- Resuspension buffer (see Recipes)
- Substrate addition (see Recipes)
- PLATE mixture (see Recipes)
Equipment
- Tube rack (any brand)
- Orbital shaker, with temperature and velocity control (e.g., Eppendorf, New Brunswick Scientific, model: Innova 44 )
- Incubator for 28-30 °C incubation of yeast cells (any brand, e.g., VWR International)
- Centrifuge with swinging rotor for 50 ml tubes (Beckman Coulter, model: Allegra 25R )
Procedure
- Sensor design
For yeast expression, CHL1/NRT1.1 or PTRs coding regions were inserted by Gateway LR reactions into the E.coli/yeast expression vectors pDRFlip30, a destination vector that sandwiches the sensory domain between Aphrodite t9 and mCerulean (Jones et al., 2014), following manufacturer’s instructions. The E. coli/yeast vector pDRFlip30 is used for expressing the NiTracs or PepTracs from a PMA1 (yeast proton ATPase) promoter fragment. pDRFlip30 contains the ADH (alcohol dehydrogenase) terminator, and the URA3 marker for auxotrophy selection in yeast (Figure 1). pDRFlip30 is a vector that allows us to sandwich the transporter of interest by translational fusion between an N-terminal Aphrodite t9 (AFPt9) variant (aphrodite is a codon-diversified gene producing Venus; Deuschle et al., 2006), lacking nine amino acids at its C-terminus and a C-terminal monomeric Cerulean (mCer; Rizzo et al., 2006). - Yeast transformation
The protease-deficient yeast strain BJ5465 [MATa ura352 trp1 leu2Δ1 his3Δ 200 pep4::HIS3 prb1Δ1.6R can1 GAL] is transformed with the pDRFlip30 vector containing the desired NiTracs and PepTracs by using the modified Lithium Acetate method from Gietz et al., 1992. In brief:- Inoculate cultures in YPD medium and grow at 30 °C overnight to OD600nm ~ 0.5.
- Spin down (2,000 x g) 1 ml of cells in microfuge tube (15 sec) for each transformation.
- Decant the supernatant and resuspend the cells in 100 μl of liquid medium by vortexing.
- Add 2 μl of 10 mg/ml carrier DNA, vortex.
- Add ~1 μg plasmid, vortex.
- Add 20 μl 1 M DTT, vortex.
- Add 0.5 ml of ‘PLATE mixture’ (100 ml stock containing 90 ml of 45% PEG4000, 10 ml of 1 M lithium acetate, 1 ml of 1 M Tris-Cl (pH 7.5), 0.2 ml of 0.5 M EDTA), vortex.
- Incubate at RT for 6-8 h or overnight.
- Heat-shock cells for 10 min at 42 °C.
- Place pipet tip directly into bottom of tube, withdraw 50-100 μl of cells and plate cells on solid -ura DropOut medium. Plates are wrapped with plastic cling wrap to prevent dehydration.
- Plates are incubated (lid down) at 30 °C for 2-3 days.
- Inoculate cultures in YPD medium and grow at 30 °C overnight to OD600nm ~ 0.5.
- Detection of NiTrac and PepTrac responses in yeast using a fluorimeter
- Single colonies are picked using sterile pipette tips and grown in a 50 ml tube containing 10 ml -ura DropOut liquid medium. Pick at least three independent colonies. Use fresh transformation; do not keep colonies for more than one week on plates to avoid mutations in yeast or plasmid.
- Place tubes in a rack and incubate in an incubator for ~15 h under agitation (230 rpm) at 30 °C until the culture reaches OD600nm ~0.5.
- Liquid cultures are subcultured after dilution to OD600nm 0.01 in the same liquid medium and grown at 30 °C until OD600nm reaches ~0.2.
- Collect the cells by centrifugation at 4,000 x g, RT for 7 min, to sediment the cells.
- Discard the supernatant and resuspend the sediment by vortexing in 10 ml ‘Wash buffer’, 15 sec, RT.
- Centrifuge as described above (step B4).
- Wash the sediment two more times as in step B4 to B6, to remove traces of growth medium.
- Resuspend the sediment to OD600nm ~0.5 in ‘Resuspension buffer’.
- Mix cells well and aliquot 100 µl of the culture into wells of a 96-well flat bottom plate.
- Fluorescence is measured in a fluorescence plate reader, in bottom reading mode using 7.5 nm bandwidth for both excitation and emission. Typically, emission spectra are recorded with the following instrument settings: λem 470-570 nm for donor (mCer), step size 5 nm, gain: 75; and λem 520-570 nm for AFPt9, step size 5 nm, gain: 75. Fluorescence from cultures harboring pDRFlip30 (donor: mCer) is measured with excitation at λexc 428 nm; AFPt9 is measured with excitation at λexc 500 nm.
- A single- or multichannel pipette is used to add 100 µl of the culture to wells (mix by pipetting up and down) and to add analyte solution to the cells. Set up at least three replicates per treatment. Try to add equal amounts of solutions to reduce variability and use well-calibrated pipettes since the assays are quantitative and sensitive to differences in volumes/ concentration of sensor and analyte.
- Record the fluorescence immediately (as fast as possible) after substrate or control solution addition. It takes about 10 min to read a full 96 well plate with the parameters mentioned above. For highly accurate analyses, measure only a few wells at a time to reduce differences in analysis time. It is also possible to use instruments with injectors that allow for immediate recording; use rapid switching between wells to record over time.
- Single colonies are picked using sterile pipette tips and grown in a 50 ml tube containing 10 ml -ura DropOut liquid medium. Pick at least three independent colonies. Use fresh transformation; do not keep colonies for more than one week on plates to avoid mutations in yeast or plasmid.
Data analysis
Subtract background fluorescence of yeast (using cells transformed with vector only) from all fluorescence values (for both spectra as well as single point measurements).
For NiTracs
NiTracs expressed in yeast respond to nitrate addition by decreasing fluorescence intensity of donor and acceptor emission (obtained with excitation at 428nm). Aphrodite-t9 emission was unaffected and served as a control or reference for normalization (obtained at 500nm excitation Figure 2A & inset). Nitrate addition (5 mM) induced a reduction in the emission spectrum, while emission of the acceptor after direct excitation of the acceptor did not change (Figure 2B). Since the Aphrodite-t9 emission is unaffected by nitrate when excited directly, Aphrodite-t9 emission can be used as a control and for normalization by using ratios instead of absolute values to compare between different cultures (e.g. mutants) (peak fluorescence intensity of Aphrodite-t9 excited at 500 nm over every point in the emission spectrum obtained with excitation at 428 nm).
For PepTracs
PepTracs respond to dipeptide by decreasing fluorescence intensity of donor and acceptor emissions (PepTrac1, PepTrac2, and PepTrac5) or by a ratio change (Aphrodite-t9 emission intensity/mCer emission intensity obtained with excitation at 428nm) in the case of PepTrac4 (Figure 3). For PepTrac1, PepTrac2, and PepTrac5, Aphrodite-t9 emission when excited at 500nm was unaffected by peptide addition in PepTracs (Figure 3A, B, & C, insect). The emission ratio change induced by addition of dipeptide for PepTrac4 is shown in Figure 2D).
Representative data
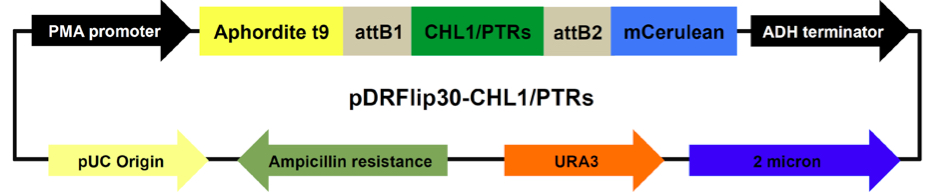
Figure 1. Map of pDRFlip30-CHL1/PTRs plasmids. Main components and their sizes (base pair, bp): PMA promoter fragment 452 bp, ADH terminator 333 bp, 2 micron replication origin 1165 bp, URA3 804 bp, Ampicillin 1863 bp, pUC origin 654 bp, CHL1 1770 bp, PTRs: PTR1 1710 bp, PTR2 1755 bp, PTR4 1635 bp, PTR5 1710 bp, Aphrodite t9 688 bp, and mCerulean 714 bp.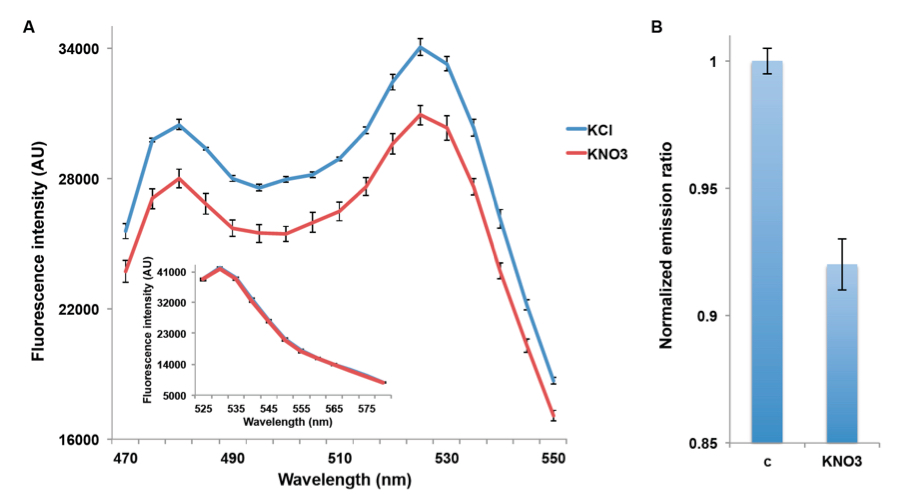
Figure 2. Decrease in emission intensity for NiTrac1 expressed in yeast cells. A. Excitation at 428 nm: addition of 5 mM potassium nitrate (red; control 5 mM KCl, blue), led to a reduction in fluorescence intensity of donor and acceptor emission. Inset: emission intensity of Aphrodite-t9 in NiTrac1 when excited at 500 nm. Inset: Aphrodite-t9 emission was unaffected. AU: arbitrary units); B. Nitrate triggers a decrease in the emission from the donor, and consequentially a reduced meission from the accepted when exciting only the donor. Nitrate-induced ratio change (peak fluorescence intensity of Aphrodite-t9 excited at 500 nm over emission spectrum at 485 nm obtained with excitation at 428 nm). Data are normalized to KCl-treated buffer (as negative control, C). The data are from the same experiment as shown in (Ho et al., 2014), but are derived from a separate analysis of independent colonies. 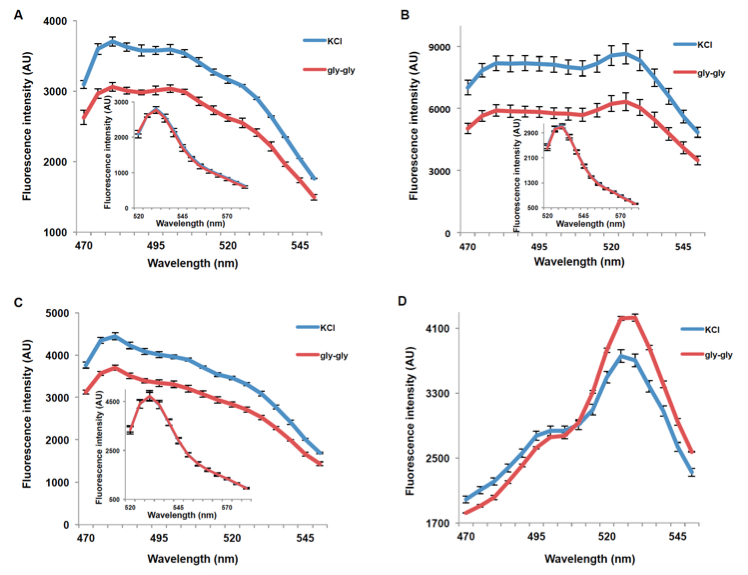
Figure 3. Fluorescence response of PepTrac1 (A), PepTrac2 (B), PepTrac5 (C), and PepTrac4 (D) expressing yeast cells. Samples were excited at 428 nm: addition of 5 mM gly-gly (red; control: 5mM KCl, blue), led to a reduction of fluorescence intensity of donor and acceptor emission in the case of PepTrac1, 2, and 5. Inset: emission of Aphrodite-t9 in PepTracs when excited at 500 nm. Aphrodite t9 emission was unaffected in PepTrac1, 2, and 5. (D) Fluorescence ratio (excitation 428 nm; emission ratio 530nm/428 nm; corresponding to mCer and Aphrodite-t9 emission, respectively) for PepTrac4 before and after addition of 5mM gly-gly dipeptide. The data are from the same experiment as shown in (Ho et al., 2014), but are derived from a separate analysis of independent colonies.
Recipes
- -ura DropOut medium
0.23 g/L -ura DropOut
1.7 g/L yeast nitrogen base w/o amino acids w/o ammonium sulfate
40% sterile filtrated glucose
Autoclave, 121 °C, 15 psi, 15 min
For liquid medium, when hand-warm, add glucose from 40% sterile filtrated stock to a final concentration of 2% under sterile hood (e.g., biosafety cabinet)
For solid medium, add 20 g/L agar before autoclaving. Add sterile filtrated glucose from 40% stock to a final concentration of 2% when medium is hand-warm before pouring plates
Adjust the pH of the -ura DropOut medium to pH 5.8 with NaOH before addition of agar and autoclaving - Wash buffer
50 mM MES buffer, adjust to pH 5.5 with NaOH - Resuspension buffer
Add agarose to final concentration 0.05% in wash buffer (Recipe 2), and then, microwave until the agarose dissolves completely and wait until medium cools to room temperature to delay sedimentation of the cells during the measurement. - Substrate addition
Add KNO3 or KCl from 1 M stock to resuspension buffer (Recipe 3) to generate analyte concentration for measurement - PLATE mixture
100 ml stock
90 ml of 45% PEG4000
10 ml of 1 M lithium acetate
1 ml of 1 M Tris-Cl (pH 7.5)
0.2 ml of 0.5 M EDTA
Acknowledgments
Methods were adapted from (Ho et al., 2014). Techniques were also adapted from other references as cited. This work has been supported by grants MCB-1021677 and MCB-1413254 from the National Science Foundation (to WBF).
References
- Deuschle, K., Chaudhuri, B., Okumoto, S., Lager, I., Lalonde, S. and Frommer, W. B. (2006). Rapid metabolism of glucose detected with FRET glucose nanosensors in epidermal cells and intact roots of Arabidopsis RNA-silencing mutants. Plant Cell 18(9): 2314-2325.
- Gietz, D., St Jean, A., Woods, R. A. and Schiestl, R. H. (1992). Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20(6): 1425.
- Ho, C. H. and Frommer, W. B. (2014). Fluorescent sensors for activity and regulation of the nitrate transceptor CHL1/NRT1.1 and oligopeptide transporters. Elife 3: e01917.
- Ho, C. H., Lin, S. H., Hu, H. C. and Tsay, Y. F. (2009). CHL1 functions as a nitrate sensor in plants. Cell 138(6): 1184-1194.
- Jones, A. M., Danielson, J. A., Manojkumar, S. N., Lanquar, V., Grossmann, G. and Frommer, W. B. (2014). Abscisic acid dynamics in roots detected with genetically encoded FRET sensors. Elife 3: e01741.
- Komarova, N. Y., Meier, S., Meier, A., Grotemeyer, M. S. and Rentsch, D. (2012). Determinants for Arabidopsis peptide transporter targeting to the tonoplast or plasma membrane. Traffic 13(8): 1090-1105.
- Leran, S., Varala, K., Boyer, J. C., Chiurazzi, M., Crawford, N., Daniel-Vedele, F., David, L., Dickstein, R., Fernandez, E., Forde, B., Gassmann, W., Geiger, D., Gojon, A., Gong, J. M., Halkier, B. A., Harris, J. M., Hedrich, R., Limami, A. M., Rentsch, D., Seo, M., Tsay, Y. F., Zhang, M., Coruzzi, G. and Lacombe, B. (2014). A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci 19(1): 5-9.
- Liu, K. H. and Tsay, Y. F. (2003). Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J 22(5): 1005-1013.
- Rizzo, M. A., Springer, G., Segawa, K., Zipfel, W. R. and Piston, D. W. (2006). Optimization of pairings and detection conditions for measurement of FRET between cyan and yellow fluorescent proteins. Microsc Microanal 12(3): 238-254.
- Tsay, Y. F., Chiu, C. C., Tsai, C. B., Ho, C. H. and Hsu, P. K. (2007). Nitrate transporters and peptide transporters. FEBS Lett 581(12): 2290-2300.
Article Information
Copyright
Ho and Frommer. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Ho, C. H. and Frommer, W. B. (2016). Design and Functional Analysis of Fluorescent Nitrate and Peptide Transporter Activity Sensors in Yeast Cultures. Bio-protocol 6(3): e1728. DOI: 10.21769/BioProtoc.1728.
- Ho, C. H. and Frommer, W. B. (2014). Fluorescent sensors for activity and regulation of the nitrate transceptor CHL1/NRT1.1 and oligopeptide transporters. Elife 3: e01917.
Category
Microbiology > Heterologous expression system > Saccharomyces cerevisiae
Cell Biology > Cell imaging > Live-cell imaging
Cell Biology > Cell imaging > Fluorescence
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









