- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantification of Ethylene Production in Tomato Leaves Infected by Xanthomonas euvesicatoria
Published: Vol 6, Iss 3, Feb 5, 2016 DOI: 10.21769/BioProtoc.1723 Views: 11362
Reviewed by: Zhaohui LiuMalou FraitureDaniel F. CaddellArsalan Daudi

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Bacterial Pathogen-mediated Suppression of Host Trafficking to Lysosomes: Fluorescence Microscopy-based DQ-Red BSA Analysis
Mădălina Mocăniță [...] Vanessa M. D'Costa
Mar 5, 2024 2902 Views

Purification of Native Dentilisin Complex from Treponema denticola by Preparative Continuous Polyacrylamide Gel Electrophoresis and Functional Analysis by Gelatin Zymography
Pachiyappan Kamarajan [...] Yvonne L. Kapila
Apr 5, 2024 2096 Views

In Silico Prediction and In Vitro Validation of Bacterial Interactions in the Plant Rhizosphere Using a Synthetic Bacterial Community
Arijit Mukherjee [...] Sanjay Swarup
Nov 5, 2025 1671 Views
Abstract
Ethylene is a gaseous plant hormone controlling fruit ripening, flower opening, leaf senescence as well as abscission, and disease symptom development. Ethylene plays a critical role in the bacterial pathogen Xanthomonas euvesicatoria (X. euvesicatoria)-elicited symptom development in tomato. This protocol describes the measurement of ethylene gas produced by tomato leaves infected with X. euvesicatoria. Infected leaflets are placed in a glass tube for 30 min without sealing. The glass tubes are then capped with a septa stopper, and incubated for an hour. A 1 ml gas sample is removed from the tube using a syringe and then injected into a gas chromatograph to quantify ethylene gas levels. This protocol will be applicable for other plants with other pathogens with modifications.
Materials and Reagents
- 1% Ethylene gas can (SCOTT Specialty Gases, catalog number: 01-04-855 )
- 1 L Tedlar® PLV Gas Sampling Bag w/Thermogreen® LB-2 Septa (Sigma-Aldrich, Supelco, catalog number: 24633 )
- 1.5 ml Microtubes (Corning, Axygen®, catalog number: MTC-150-C )
- 1 ml Syringes Without Needles (BD, catalog number: 309659 )
- 25 G Needle (BD, catalog number: 305122 )
- 15 ml Polypropylene Centrifuge Tubes (Greiner Bio-One GmbH, catalog number: 188271 )
- 16 mm diameter x 100 mm Glass tubes (VWR International, catalog number: 47729-576 )
- 18 mm diameter x 150 mm Glass tubes (VWR International, catalog number: 47729-583 )
- Suba-Seal® septa (Sigma-Aldrich, catalog number: Z124613 )
- Tomato plants (4-5 week-old) were grown in a greenhouse or growth chamber (16 h light, 25~28 °C)
- Xanthomonas euvesicatoria (Xcv) strain 85-10 wild type and mutants (for example, type III effector deletion mutants)
Note: Wild type strain is available upon request. - Magnesium chloride hexahydrate (Sigma-Aldrich, catalog number: M2393 )
- Peptone (BD, catalog number: 211677 )
- Yeast extract (BD, catalog number: 212750 )
- Agar (BD, catalog number: 214530 )
- Glycerol (Certified ACS) (Fisher Chemical, catalog number: G334 )
- Sodium hydroxide (Sigma-Aldrich, catalog number: 221465 )
- Distilled water
- NYGA medium (see Recipes)
- 10 mM MgCl2 (see Recipes)
Equipment
- 28 ºC Incubator (VWR International, catalog number: 414005-128 )
- Vortexer (Scientific Indrustries, model: Vortex-Genie 2 )
- Spectrophotometer (Amersham Biosciences, model: Ultrospec 3100 Pro )
- Gas chromatograph (GC) (Shimadzu Corporation, model: GC-8A )
Procedure
- Bacterial inoculum preparation and inoculation
- Streak out each Xcv strain from -80 °C glycerol stocks using a sterilized toothpick onto an independent sterile NYGA agar plate with appropriate antibiotics and grow for 2 d in a 28 °C incubator.
- For each strain analyzed, suspend a portion of the bacterial cells from the agar plate using a sterilized toothpick or pipet tip in 1 ml of 10 mM MgCl2 in a 1.5 ml microtube. Vortex the cell suspension well and check its optical density at 600 nm (OD600) by using a spectrophotometer. To prepare the inoculum, dilute the cell suspension with 10 mM MgCl2 to obtain an OD600 = 0.2. (Different inoculum concentrations should be empirically tested for optimal ethylene production.)
- Inoculate each leaflet of middle leaves with 10 mM MgCl2 (mock inoculation) or a bacterial inoculum using a 1 ml needleless syringe (Figure 1). Carefully remove excess solution on the surface of the leaflet with tissue paper. Use leaflets on the same branch for comparison to control for leaf age. For statistical analysis, prepare a minimum of three biological replicates.
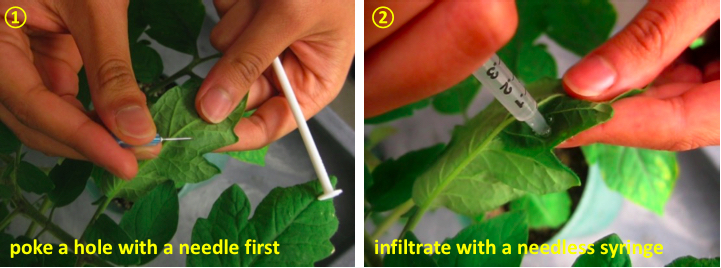
Figure 1. Hand infiltration with needless syringe - Move the inoculated plants into a plant growth chamber or greenhouse (~ 25-28 °C, 16 h day/8 h night cycle).
- Streak out each Xcv strain from -80 °C glycerol stocks using a sterilized toothpick onto an independent sterile NYGA agar plate with appropriate antibiotics and grow for 2 d in a 28 °C incubator.
- Ethylene gas measurement
- At 0, 2, and 3 days post-inoculation (dpi), excise the leaflets (mock or Xcv-infected) and measure their weight.
- Roll each leaflet and then place it into a glass tube (16 mm diameter x 100 mm) for 30 min without sealing.
- Then cap each glass tube with a Suba-Seal Septa stopper (Figure 2).
Note: It is important to use the smallest size glass tube to reduce the volume of the gas space to concentrate the ethylene emitted by each leaflet.
Figure 2. Rolled leaflet in a glass tube capped with a septa stopper - Incubate the glass tubes for 1 h at room temperature.
- Using a 25 gauge needle attached to a 1 ml syringe, remove 1 ml of the gas sample from each glass tube (Figure 3) and inject it into a gas chromatograph.
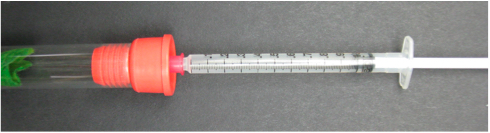
Figure 3. Gas sampling from a glass tube capped with a septa stopper - Use company instruction manual to detect ethylene [e.g., Shimadzu's GC instruction manual (https://store.shimadzu.com/p-52629-instruction-manualgc-8apfgc-8a.aspx)].
- At 0, 2, and 3 days post-inoculation (dpi), excise the leaflets (mock or Xcv-infected) and measure their weight.
- Measuring ethylene using a standard curve
- Use the needle attachment on the 1% ethylene can to pump ethylene gas into a Tedlar bag. Label bag as standard 1. Final concentration of standard 1 = 1% = 10,000 nl of ethylene/ml.
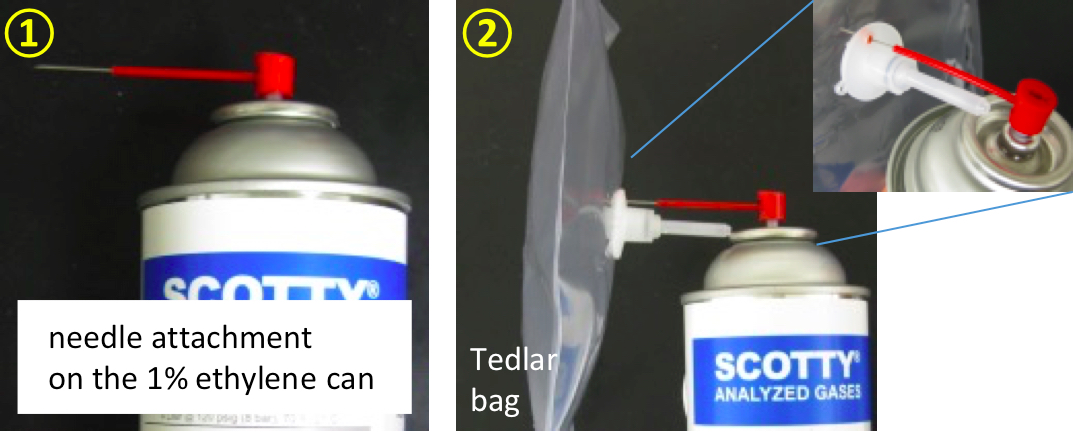
Figure 4. Pumping ethylene gas into a Tedlar bag - Take 1 ml of 1% ethylene from the ethylene-filled Tedlar bag and inject it into a capped glass tube (18 mm diameter x 150 mm glass tube with Suba-seal septa). Label tube as standard 2. Final concentration of standard 2 = 0.0352% or 352 nl of ethylene/ml. (An empty capped tube contains 27.4 ml of air. Ethylene concentration in standard 2 = 1% /1 ml + 27.4 ml.)
- Take 1 ml from capped glass tube (standard 2; 18 mm diameter x 150 mm glass tube with Suba-seal septa) and inject it into a capped tube labeled standard 3 (18 mm diameter x 150 mm glass tube with Suba-seal septa). Final concentration of standard 3 = 0.00124% or 12.4 nl of ethylene/ml.
- Inject 1 ml of each standard on the GC and determine the peak area.
- Construct a standard curve by plotting the peak area (X-axis) by nl of ethylene (Y-axis).
- Using the standard curve, determine the level of ethylene in each inoculated leaflet. Ethylene levels are defined as nl of ethylene/gram of fresh weight/hour. Representative results can be found in Lund et al. (1998) (Figure 1C and Figure 2C) and Kim et al. (2013) (Figure 1A-B).
- Use the needle attachment on the 1% ethylene can to pump ethylene gas into a Tedlar bag. Label bag as standard 1. Final concentration of standard 1 = 1% = 10,000 nl of ethylene/ml.
Recipes
- NYGA medium (1 L)
Peptone 5 g
Yeast extract 3 g
Agar 15 g
Glycerol 20 ml
Add distilled water to make up 1 L
Adjust pH at 7.0 with 1 N sodium hydroxide
Sterilize the medium by autoclaving for 20 min - 10 mM MgCl2
Magnesium chloride hexahydrate 2.03 g
Add distilled water to make up 1 L
Sterilize the medium by autoclaving for 20 min
Acknowledgments
This protocol is adopted from Lund et al. (1998). W. Stork was supported by United States Department of Agriculture NIFA Grant 2012-67011-19669. M. B. Mudgett was supported by National Institutes of Health Grant 2 R01 GM068886-06A1.
References
- Kim, J. G., Stork, W. and Mudgett, M. B. (2013). Xanthomonas type III effector XopD desumoylates tomato transcription factor SlERF4 to suppress ethylene responses and promote pathogen growth. Cell Host Microbe 13(2): 143-154.
- Lund, S. T., Stall, R. E. and Klee, H. J. (1998). Ethylene regulates the susceptible response to pathogen infection in tomato. Plant Cell 10(3): 371-382.
- Stork, W., Kim, J. G. and Mudgett, M. B. (2015). Functional analysis of plant defense suppression and activation by the Xanthomonas core Type III effector XopX. Mol Plant Microbe Interact 28(2): 180-194.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Kim, J., Stork, W. and Mudgett, M. B. (2016). Quantification of Ethylene Production in Tomato Leaves Infected by Xanthomonas euvesicatoria. Bio-protocol 6(3): e1723. DOI: 10.21769/BioProtoc.1723.
Category
Plant Science > Plant immunity > Disease bioassay
Microbiology > Microbe-host interactions > Bacterium
Biochemistry > Other compound > Plant hormone > Ethylene
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









