- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Preparation of Chloroplast Lipid Membrane and Lipid-protein Interaction Assay
Published: Vol 6, Iss 3, Feb 5, 2016 DOI: 10.21769/BioProtoc.1720 Views: 10745
Reviewed by: Tie LiuVenkatasalam ShanmugabalajiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

GC-MS-Based Analysis of Methanol: Chloroform-extracted Fatty Acids from Plant Tissues
Manish Kumar Patel [...] Jitendra Kumar Thakur
Sep 20, 2018 15450 Views

Sorghum bicolor Extracellular Vesicle Isolation, Labeling, and Correlative Light and Electron Microscopy
Deji Adekanye [...] Jeffrey L. Caplan
Oct 5, 2024 2118 Views

PhosphoLIMBO: An Easy and Efficient Protocol to Separate and Analyze Phospholipids by HPTLC From Plant Material
Louise Fougère [...] Yohann Boutté
Sep 5, 2025 1344 Views
Abstract
Lipid-Protein interaction assay is a method to search lipids, which are bound with proteins in vitro. Since membranes that are spotted with chloroplast lipids such as monogalactosyldiacylglycerol (MGDG), digalactosyldiacylglycerol (DGDG), and sulfoquinovosyldiacylglycerol (SQDG) are not commercially available, we extracted these lipids from cyanobacterial cells and spotted them onto membranes. The prepared membranes could be used for lipid-protein interaction assay.
Keywords: Lipid-protein interaction assayMaterials and Reagents
- Preparation of lipids for chloroplast lipid membrane
- Amersham Protran Premium NC0.2 (GE Healthcare, Dharmacon, catalog number: AP-10600081 )
- TLC silica gel 60 plate (Merck Millipore Corporation, catalog number: 105721 )
- Fused silica capillary column, 0.25 mm x 50 m (Shinwa Chemical Industries, catalog number: HR-SS-10 )
- Cells of the cyanobacterium, Synechocystis sp. PCC 6803 (for lipid extraction)
- Methanol (Wako Pure Chemical Industries, Siyaku, catalog number: 137-01823 )
- Chloroform (Wako Pure Chemical Industries, Siyaku, catalog number: 038-02601 )
- 0.01% primuline (Sigma-Aldrich, catalog number: 206865 ), in 80% acetone
- Pentadecanoic acid (Sigma-Aldrich, catalog number: 91446-5G )
- Methanolic HCl (Sigma-Aldrich, catalog number: 33050-U ), dilute with methanol 3 times
- Hexane (Wako Pure Chemical Industries, Siyaku, catalog number: 085-00411 )
- Amersham Protran Premium NC0.2 (GE Healthcare, Dharmacon, catalog number: AP-10600081 )
- Lipid-Protein interaction assay
- PIP Strips, Membrane Lipid Strips (Echelon Biosciences, catalog numbers: P-6001 and P-6002 )
- Hybri-Bag (Cosmo Bio, catalog number: S-1001 )
- Albumin from bovine serum (BSA), fatty acid free (Wako Pure Chemical Industries, Siyaku, catalog number: 017-15146 )
- Purified tagged proteins
- An anti-penta-His mouse monoclonal antibody (QIAGEN, catalog number: 34660 ) and an anti-GST mouse monoclonal antibody (Sigma-Aldrich, catalog number: G1160 )
- Horseradish peroxidase (HRP)-conjugated goat anti-mouse secondary antibody (Thermo Fisher Scientific, catalog number: 31432 )
- HRP substrate solution (Thermo Fisher Scientific, Pierce, catalog number: NCI32132 )
- Phosphate (pH 7.4)
- NaCl
- 0.1 % Tween-20
- Phosphate buffered saline (PBS) (see Recipes)
- PBST (see Recipes)
- PIP Strips, Membrane Lipid Strips (Echelon Biosciences, catalog numbers: P-6001 and P-6002 )
Equipment
- Glass tubes (Iwaki, catalog numbers: 71-088-004 and 71-063-006 )
- Pasteur pipette (Sansyo, Iwaki, catalog number: IK-PAS-9P )
- Spectrophotometer (Scinteck Instruments, Pharmacia Biotech, model: Ultrospec 2000 )
- Vortex mixer
- Centrifuge (TOMY SEIKO CO., model: LC-120 )
- Vacuum dryer (Shimadzu Corporation, model: SPE-200 centrifuge evaporator )
- TLC developing chamber (AS ONE, catalog number: CK-0544-060 )
- Hair dryer (commercially available)
- UV illuminator
- Razor blade
- Heat block
- Gas chromatograph (Shimadzu Corporation, model: GC-18A )
- Fused silica capillary column, 0.25 mm x 50 m (Shinwa Chemical Industries, catalog number: HR-SS-10 )
- Data processor (Shimadzu Corporation, model: C-R2AX )
Procedure
- Preparation of chloroplast lipid membrane
- Collect cyanobacterial cells by centrifugation and suspend the collected cells in 2 ml of 0.9% KCl. 3 x 109 cells of Synechocystis PCC 6803 are sufficient [Number of cells were determined by spectrophotometer. Optical density at 730 nm (OD730) = 1.0 corresponds to 1 x 108 cells /ml for Synechocystis PCC 6803].
- Add 7.5 ml of methanol:chloroform (2:1, v/v) and vortex for 1 min.
- Add 2.5 ml of chloroform and 2.5 ml of H2O, and vortex.
- Centrifuge at 700 x g for 10 min. Collect lower phase, put it into clean tube and dry in a vacuum dryer. Measure the weight of the tube before use and after drying lipids. Calculate the yield of lipids.
- Dissolve dried lipids in chloroform at 10 mg/ml.
- Add developing solvent into a TLC developing chamber, close and leave it for 30 min as shown in Figure 1. Developing solvent is acetone:benzene:ultrapure water = 91:30:8 (v/v/v). Spot the chloroform solution containing 1-3 mg of lipids onto a TLC plate and place the plate into the TLC developing chamber.
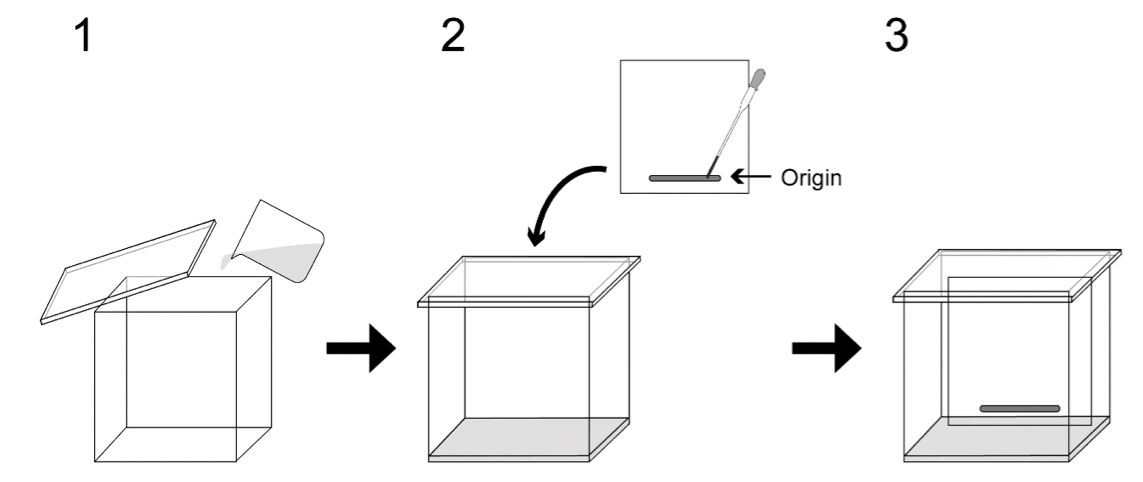
Figure 1. Separation of lipids on TLC plate. 1. Add developing solvent into a TLC developing chamber; 2. Spot lipid solution onto a TLC plate and place the plate into the chamber; 3. The TLC plate is developed in the chamber to separate lipids. - When the solvent front has reached 1 cm from the top of the plate, remove the plate from the chamber and dry with cool wind provided by hair dryer.
- Spray 0.01% primuline in 80% acetone onto the plate. Lipids are viewed as spots under long wavelength of UV light as shown in Figure 2. Mark the corresponding position of each lipid and scrape the silica gel in the each spot from the TLC plate that is grass plate coated with silica gel. The silica gel can be scraped from the plate with a razor blade.
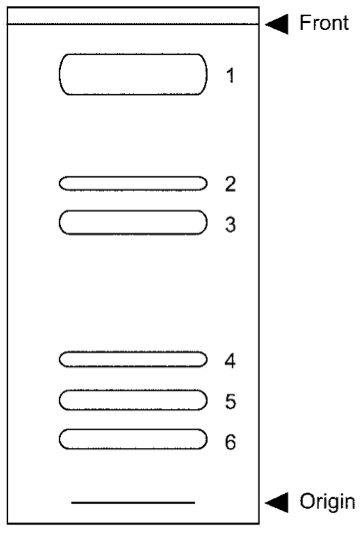
Figure 2. Thin-layer chromatogram of cyanobacterial lipids visualized by UV. Total lipids extracted from cyanobacteria were developed in acetone:benzene:ultrapure water=91:30:8. 1. Pigments; 2. Monoglucosyldiacyalglycerol; 3. Monogalactosyldiacylglycerol (MGDG); 4. Digalactosyldiacylglycerol (DGDG); 5. Sulfoquinovosyldiacylglycerol (SQDG); 6. Phosphatidylglycerol (PG). - Add 1 ml of chloroform to the silica gel scraped from the plate and vortex. Spin down the silica gel and transfer supernatant to a new tube. Repeat this extraction 3 times and evaporate chloroform in a vacuum dryer. Dissolve the obtained lipids in 100 μl of chloroform. The following 10 to 12 steps are required to determine the yield of obtained lipids.
- Take 5 μl of the chloroform solution containing each lipid to a new tube with a screw cap and add 50 nmol of pentadecanoic acid as an internal standard and 2.5 ml of 3% (w/v) methanolic HCl. Close the tube with the screw cap and incubate at 80 °C for 3 h. During the incubation fatty acids bound to lipids are derivatized to fatty acid methyl esters.
- After cooling the tube to room temperature, add 2.5 ml of hexane and vortex vigorously. Fatty acid methyl esters are extracted with hexane. Leave to stand for 5 min and collect upper phase (hexane phase), put it into a clean tube and evaporate hexane in a vacuum dryer.
- Dissolve the obtained fatty acid methyl esters in 50 μl of hexane and analyze them with a gas chromatograph. Temperatures for the injector, column and detector chambers were set at 250 °C, 170 °C and 250 °C, respectively. Determine lipid content by calculating total amount of fatty acid methyl esters based on the peak areas of fatty acid methyl esters in the obtained chromatograms. Pentadecanoic acid is not contained in cyanobacterial lipids, thus it can be used as an internal standard.
- Dilute lipids (the obtained lipids at the step A9) with chloroform to 0.1, 1 and 10 mg/ml.
- Spot the lipids corresponding to 0.1, 1 and 10 mg/ml in 1 μl of chloroform onto Amersham Protran Premium membrane and dry completely.
- Collect cyanobacterial cells by centrifugation and suspend the collected cells in 2 ml of 0.9% KCl. 3 x 109 cells of Synechocystis PCC 6803 are sufficient [Number of cells were determined by spectrophotometer. Optical density at 730 nm (OD730) = 1.0 corresponds to 1 x 108 cells /ml for Synechocystis PCC 6803].
- Lipid-protein interaction assay
- Incubate the membrane with 5 ml of blocking solution (3% fatty acid-free BSA in PBS) for 1 h at room temperature with gentle agitation.
- Discard the blocking solution and incubate the membrane with 2 ml of blocking solution containing 0.5 μg/ml tagged-protein (His-tagged or GST-tagged protein) overnight at 4 °C. Put the membrane and solution into a Hybri-Bag and seal it.
- Discard the protein solution and wash the membrane with 10 ml of PBST three times at room temperature with gentle agitation for 10 min each.
- Discard the solution and incubate the membrane with 2 ml of blocking solution containing primary antibody (anti-penta-His mouse antibody 1:2,000 or anti-GST mouse monoclonal antibody 1:2,000) for 1 h at room temperature with gentle agitation. Put the membrane and solution into a Hybri-Bag and seal it.
- Discard the solution and wash the membrane with 10 ml of PBST three times at room temperature with gentle agitation for 10 min each.
- Discard the solution and incubate the membrane with 2 ml of blocking solution containing secondary antibody (HRP-conjugated goat anti-mouse antibody 1:20,000) for 1 h at room temperature with gentle agitation. Put the membrane and solution into a Hybri-Bag and seal it.
- Discard the solution and wash the membrane with 10 ml of PBST three times at room temperature with gentle agitation for 10 min each.
- Incubate the membrane with enough HRP substrate solution to cover the membrane (2 to 3 ml) at room temperature for 5 min.
- Wash the membrane with ultrapure water to stop the reaction and dry the membrane.
- Scan the membrane to record the obtained data.
- Incubate the membrane with 5 ml of blocking solution (3% fatty acid-free BSA in PBS) for 1 h at room temperature with gentle agitation.
Representative data

Figure 3. Representative results of Lipid-Protein Interaction Assay. Each spot on the left (PIP Strip) and middle membrane (Membrane Lipid Strip) contained 100 pmol of lipids. The right membrane contained chloroplast lipids, MGDG, DGDG, SQDG, and PG, and also contained cardiolipin (CL) as the control. The membranes were incubated with 0.5 μg/mL GST-PDV2. The abbreviations used are: LPA, lysophosphatidic acid; LPC, lysophosphatidylcholine; PI, phosphatidylinositol; PI3P, phosphatidylinositol 3-phosphate; PI4P, phosphatidylinositol 4-phosphate; PI5P, phosphatidylinositol 5-phosphate; PE, phosphatidylethanolamine; PC, phosphatidylcholine; PI(3, 4)P2, phosphatidylinositol 3, 4-bisphosphate; PI(3, 5)P2, phosphatidylinositol 3, 5-bisphosphate; PI(4, 5)P2, phosphatidylinositol 4, 5-bisphosphate; S1P, sphingosine 1-phosphate; PI(3, 4, 5)P3, phosphatidylinositol 3, 4, 5-triphosphate; PA, phosphatidic acid; PS, phosphatidylserine; TAG, triacylglycerol; DAG, 1, 2-diacylglycerol. The image was reprinted from Reference 2 with permission from The Plant Cell; copyright American Society of Plant Biologists (www.plantcell.org).
Recipes
- PBS
10 mM phosphate (pH 7.4)
150 mM NaCl - PBST
PBS
0.1 % Tween-20
Acknowledgments
This protocol was adapted from the original work (Okazaki et al., 2015) to provide the detailed procedures. This work was supported by JSPS KAKENHI Grant Number 26840089.
References
- Bligh, E. G. and Dyer, W. J. (1959). A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37(8): 911-917.
- Okazaki, K., Miyagishima, S. Y. and Wada, H. (2015). Phosphatidylinositol 4-phosphate negatively regulates chloroplast division in Arabidopsis. Plant Cell 27(3): 663-674.
- Sato, N. and Murata, N. (1988). Membrane lipids. Meth Enzymol 167: 251-259.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Okazaki, K., Miyagishima, S. and Wada, H. (2016). Preparation of Chloroplast Lipid Membrane and Lipid-protein Interaction Assay. Bio-protocol 6(3): e1720. DOI: 10.21769/BioProtoc.1720.
Category
Plant Science > Plant biochemistry > Lipid
Biochemistry > Lipid > Lipid isolation
Biochemistry > Lipid > Lipid-protein interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









