- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Measurement of PI4P Levels in Intact Chloroplasts Isolated from Arabidopsis thaliana
Published: Vol 6, Iss 3, Feb 5, 2016 DOI: 10.21769/BioProtoc.1719 Views: 10294
Reviewed by: Tie LiuYingnan HouArsalan Daudi

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Sorghum bicolor Extracellular Vesicle Isolation, Labeling, and Correlative Light and Electron Microscopy
Deji Adekanye [...] Jeffrey L. Caplan
Oct 5, 2024 2118 Views

A New Approach to Detect and Semi-quantify All Molecular Species and Classes of Anionic Phospholipids Simultaneously in Plant Samples
Manon Genva [...] Laetitia Fouillen
Apr 20, 2025 1731 Views

PhosphoLIMBO: An Easy and Efficient Protocol to Separate and Analyze Phospholipids by HPTLC From Plant Material
Louise Fougère [...] Yohann Boutté
Sep 5, 2025 1344 Views
Abstract
Phosphatidylinositol 4-phosphate (PI4P), a major species of phosphoinositides, modulates many fundamental cellular processes. We have recently revealed that PI4P plays an important role in chloroplast division as a negative regulator. Despite its importance in chloroplasts, the content of PI4P in chloroplasts is very low and it is difficult to measure PI4P levels. In this protocol, we describe a simple method that we have developed for measurement of low level of PI4P in chloroplasts. Intact chloroplasts were isolated by a basic method using Percoll gradient centrifugation and acidic lipids were extracted from the isolated chloroplasts. The extracted acidic lipids including PI4P were spotted onto the membrane strip, which had been pre-spotted with PI4P standards and other phosphoinositides as negative controls. PI4P in the spot of acidic lipids on the membrane was detected using a PI4P binding protein.
Keywords: Phosphatidylinositol 4-phosphateMaterials and Reagents
- Isolation of intact chloroplasts
- Miracloth (Merck Millipore Corporation, Calbiochem®, catalog number: 475855 )
- Gauze (Hakujuji Co., model: FC-gauze )
- Seedlings of Arabidopsis thaliana
Wild-type (Columbia-0) plants, phosphatidylinositol 4-kinase (PI4K) α1 knockdown plants, pi4kβ2-1 mutants and PI4Kα1 knockdown plants of pi4kβ2-1 mutants were grown for 4 days on MS agar plates and then transferred onto agar plates with or without 5 μM dexamethazone (DEX) and grown for 1 week. The down-regulation of PI4Kα1 expression was induced by DEX treatments in PI4Kα1 knockdown plants and PI4Kα1 knockdown plants of pi4kβ2-1 mutants. For treatments with inhibitors, wild-type 4-d-old seedlings were transferred onto agar plates with PI4K inhibitors, 200 μM wortmannin (WM) or 25 μM phenylarsine oxide (PAO), or a phosphatidylinositol 3-kinase (PI3K) inhibitor, 50 μM LY294002 (LY), or without inhibitors (DMSO) and grown for 3 days. - Percoll (GE Healthcare, Dharmacon, catalog number: 7-0891-01 )
- Bradford assay kit (Bio-Rad Laboratories, catalog number: 500-0006JA )
- 1x protease inhibitor cocktail (Nakarai, catalog number: 03969-21)
Note: Currently, it is “Nacalai tesque, catalog number: 03969-21 ”. - Sorbitol
- HEPES-KOH (pH 7.5)
- EDTA
- Grinding buffer (see Recipes)
- 80% Percoll or 40% Percoll (see Recipes)
- Miracloth (Merck Millipore Corporation, Calbiochem®, catalog number: 475855 )
- Extraction of PI4P
- 0.75 M Trichloroacetic acid (TCA) (Wako Pure Chemical Industries, Siyaku, catalog number: 203-04952 )
- 5% (w/v) TCA with 1 mM EDTA
- Methanol (Wako Pure Chemical Industries, Siyaku, catalog number: 137-01823 ): Chloroform (Wako Pure Chemical Industries, Siyaku, catalog number: 038-02601 ) (2:1, v/v)
- Methanol: chloroform: 12 N HCl (Wako Pure Chemical Industries, Siyaku, catalog number: 080-01066 ) (80:40:1, v/v/v)
- 0.75 M Trichloroacetic acid (TCA) (Wako Pure Chemical Industries, Siyaku, catalog number: 203-04952 )
- Measurement of PI4P levels
- PI(4)P Mass Strip Kit (Echelon Biosciences, catalog number: K-4000E )
- Albumin from bovine serum (BSA), fatty acid free (Wako Pure Chemical Industries, Siyaku, catalog number: 017-15146 )
- HRP substrate solution (Thermo Fisher Scientific, Pierce, catalog number: NCI32132 )
- Phosphate (pH 7.4)
- NaCl
- 0.1% (w/v) Tween-20
- Phosphate buffered saline (PBS) (see Recipes)
- PBST (see Recipes)
- PI(4)P Mass Strip Kit (Echelon Biosciences, catalog number: K-4000E )
Equipment
- Homogenizer (Microtec Co., model: NS-51 )
- Centrifuge (TOMY SEIKO CO., models: MX-200 and GX-250 )
- Swing rotor (TS-7C)
- Paintbrush
- Pasteur pipette (Sansyo, Iwaki, catalog number: IK-PAS-9P )
- Vacuum dryer (Centrifuge evaporator) (Shimadzu Corporation, model: SPE-200 )
- Sonicator (SHARP CORPORATION, model: UT-106 )
Procedure
- Isolation of intact chloroplasts
- Sampling 0.5-1 g of seedlings of Arabidopsis thaliana. Whole seedlings for one-week-old plants or shoots for eleven-day-old plants were used.
- Add 6 ml of grinding buffer and homogenize with homogenizer 3-5 times for 3 sec on ice.
- Filtrate the homogenate through 2 layers of gauze and a layer of Miracloth (put gauzes on the Miracloth).
- Centrifuge at 2,500 x g for 15 min at 4 °C. Discard the supernatant and gently resuspend the pellet in 1.5 ml of grinding buffer with a paintbrush. Brush the pellet gently with paintbrush little by little to avoid breaking of intact chloroplasts. Note that pipetting the pellet breaks chloroplasts.
- Add 1.5 ml of grinding buffer and 1 ml of 80% Percoll and gently mix. (Final concentration of Percoll is 20%.)
- Prepare Percoll step gradients. Layering first 3 ml of 80% Percoll and then 5 ml of 40% Percoll in 15 ml centrifuge tubes. Put the tip of pipette on the surface and slowly lay 40% Percoll to avoid disturbance of the gradient. See Figure 1.
- Lay 4 ml of crude chloroplast suspension (the results of step A5) on the top of Percoll gradients.
- Centrifuge at 3,200 x g for 30 min at 4 °C using a swing rotor. Set speed of acceleration and deceleration at minimum.
- Collect a green band with a Pasteur pipette corresponding to intact chloroplasts at the interface between 40% and 80% to 2 ml tubes. Remove carefully the upper band (broken chloroplasts) and transfer lower band to 2 ml tubes with a Pasteur pipette. See Figure 1.
- Add grinding buffer up to 2 ml and centrifuge at 700 x g for 5 min at 4 °C.
- Discard the supernatant and wash the pellet two times by resuspension in 1 ml of grinding solution and centrifugation at 700 x g for 5 min at 4 °C.
- Suspend the pellet (intact chloroplasts) in 0.3 ml of grinding buffer.
- Take a part of the samples for measurement of protein concentration (step B9).
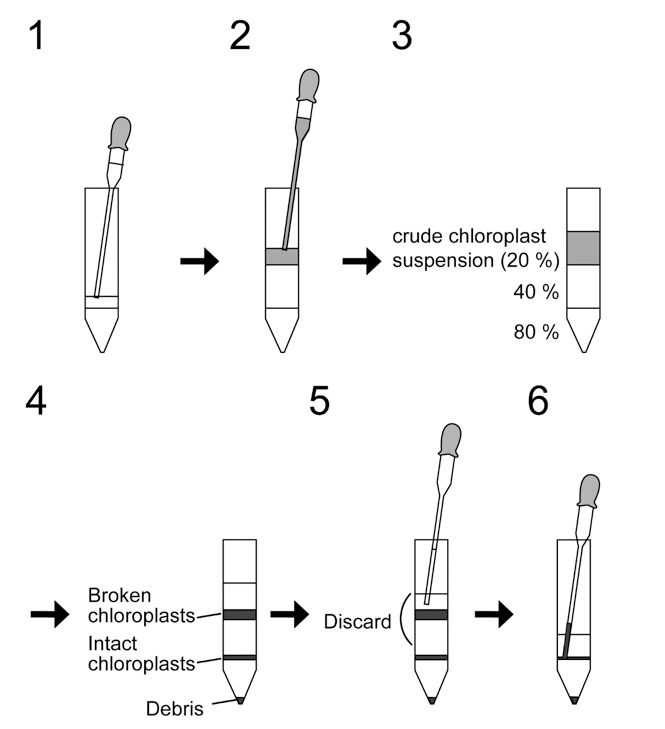
Figure 1. Isolation of intact chloroplasts with Percoll gradient centrifugation. 1. Lay 40% Percoll on 80% Percoll; 2. Lay crude chloroplast suspension on 40% Percoll; 3. The tube before centrifugation; 4. The tube after centrifugation; 5. Remove the upper band (broken chloroplasts); 6. Collect the lower band (intact chloroplasts).
- Sampling 0.5-1 g of seedlings of Arabidopsis thaliana. Whole seedlings for one-week-old plants or shoots for eleven-day-old plants were used.
- Extraction of PI4P
- Add 750 μl of ice cold 0.75 M TCA to 250 μl of intact chloroplast samples. Vortex and incubate on ice for 5 min.
- Centrifuge at 700 x g for 5 min at 4 °C and discard the supernatant. Add 1 ml of 5% TCA with 1 mM EDTA to the pellet and vortex for 30 sec.
- Centrifuge at 700 x g for 5 min at room temperature and discard the supernatant. Add 1 ml of methanol:chloroform (2:1) to the pellet for extraction of neutral lipids and vortex 3 times for 30 sec at an interval of 5 min at room temperature.
- Centrifuge at 700 x g for 5 min at room temperature and discard the supernatant. Add 1 ml of methanol:chloroform (2:1) to the pellet for extraction of neutral lipids again and vortex for 1 min at room temperature.
- Centrifuge at 700 x g for 5 min at room temperature and discard the supernatant. Add 750 μl of methanol:chloroform:12 N HCl (80:40:1) to the pellet for extraction of acidic lipids and vortex 4 times for 30 sec at an interval of 5 min.
- Add 250 μl of chloroform and 450 μl of 0.1 M HCl and vortex.
- Centrifuge at 700 x g for 5 min, collect the lower phase and transfer it to new tubes.
- Dry samples in a vacuum dryer.
- Measure protein concentration. Add 1-10 μl of samples (step A13) into 1 ml of Bradford assay reagents (1:5 dilution). Incubate for 5 min at room temperature and measure absorbance at 595 nm. Measure absorbance of standard protein samples too. Make a standard curve using standard protein samples and calculate the PI4P concentration according to the reads. The concentration of protein is usually 0.3-0.8 mg/ml.
- Add 750 μl of ice cold 0.75 M TCA to 250 μl of intact chloroplast samples. Vortex and incubate on ice for 5 min.
- Measurement of PI4P levels
- Add 15 μl of chloroform: methanol: water (1:2:0.8, v/v/v) to the sample. Vortex for 30 sec and sonicate (100% output) in icy water bath for 5 min. Vortex again for 30 sec.
- Centrifuge 15,000 x g for 5 min and transfer 10 μl of supernatant to new tube. Do not suck the pellet at the bottom of tube.
- Spot samples onto a PI(4)P Strip. Spot 1 μl of sample at a time. Before spotting sample again in the same area, dry the spot. After all samples are spotted, dry the strip for 30 min at room temperature. See Figure 2.
- Incubate the membrane with blocking solution (3% fatty acid-free BSA in PBS) for 1 h at room temperature with gentle agitation.
- Incubate the membrane with 5 ml of blocking solution containing 1 vial of PI(4)P Detector (2.5 μg protein) for 1 h at room temperature with gentle agitation.
- Discard the solution and wash the membrane with 10 ml of PBST three times at room temperature with gentle agitation for 5 min each.
- Incubate the washed membrane with 5 ml of blocking solution containing 75 μl of secondary detector for 1 h at room temperature with gentle agitation.
- Discard the solution and wash the membrane three times with 10 ml of PBST at room temperature with gentle agitation for 5 min each.
- Incubate the washed membrane with enough ECL solution to cover the membrane at room temperature for 5 min.
- Wash the membrane with water three times to stop the reaction and dry the membrane.
- Scan the membrane to record the obtained data. See Figure 2.
- Add 15 μl of chloroform: methanol: water (1:2:0.8, v/v/v) to the sample. Vortex for 30 sec and sonicate (100% output) in icy water bath for 5 min. Vortex again for 30 sec.
Representative data
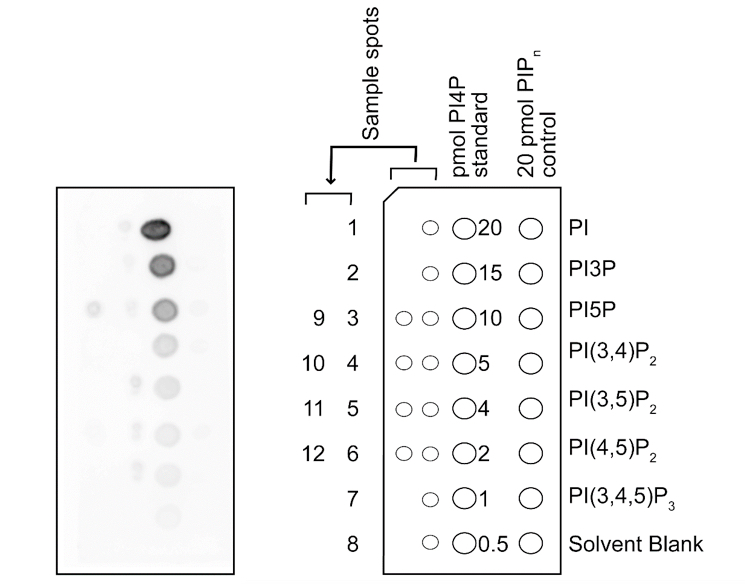
Figure 2. Representative results of PI4P detection assay. Samples extracted from intact chloroplasts were spotted left side of the PI(4)P Strip. PI4P standards (20, 15, 10, 5, 4, 2, 1, and 0.5 pmol of PI4P) and phosphoinositide controls were pre-spotted on the membrane. Lipids extracted from isolated chloroplasts of wild-type plants, phosphatidylinositol 4-kinase (PI4K) α1 knockdown plants, pi4kβ2-1 mutants and PI4Kα1 knockdown plants of pi4kβ2-1 mutants treated with/without 5 μM dexamethasone (DEX) were spotted onto the membrane. The down-regulation of PI4Kα1 expression was induced by DEX treatments in PI4Kα1 knockdown plants and PI4Kα1 knockdown plants of pi4kβ2-1 mutants. Lipids extracted from isolated chloroplasts of wild-type plants treated with/without PI4K inhibitors, 200 μM wortmannin (WM) or 25 μM phenylarsine oxide (PAO), or a phosphatidylinositol 3-kinase (PI3K) inhibitor, 50 μM LY294002 (LY), were also spotted. Sample spots are: 1. Wild-type plants without DEX; 2. Wild-type plants treated with DEX; 3. PI4Kα1 knockdown plants without DEX; 4. PI4Kα1 knockdown plants treated with DEX; 5. pi4kβ2-1 mutants without DEX; 6. pi4kβ2-1 mutants treated with DEX; 7. PI4Kα1 knockdown plants of pi4kβ2-1 mutants treated without DEX; 8. PI4Kα1 knockdown plants of pi4kβ2-1 mutants treated with DEX; 9. Wild-type plants without inhibitor; 10. Wild-type plants with WM; 11. Wild-type plants with PAO; 12. Wild-type plants with LY. The abbreviations used are: PI, phosphatidylinositol; PI3P, phosphatidylinositol 3-phosphate; PI5P, phosphatidylinositol 5-phosphate; PI(3, 4)P2, phosphatidylinositol 3, 4-bisphosphate; PI(3, 5)P2, phosphatidylinositol 3, 5-bisphosphate; PI(4, 5)P2, phosphatidylinositol 4, 5-bisphosphate; PI(3, 4, 5)P3, phosphatidylinositol 3, 4, 5-triphosphate.
Notes
- The samples and Percoll gradient tubes should be kept on ice during procedure A.
- Do not homogenize samples too much. Three to five times for 3 sec are usually enough. Longer homogenization results in lower yield of intact chloroplasts.
- After step B3, do not keep samples on ice.
- Intactness of chloroplasts can be assessed by phase contrast microscope. Intact chloroplasts are surrounded by bright halos of light. Broken chloroplasts look dark. See Figure 3 of Klinkenberg (2014).
- Extraction of neutral lipids should be done twice for removing chlorophyll.
- Do not suck the pellet at step C2. It contains left over proteins from extraction steps and makes hydrophobic film on the membrane. The hydrophobic film prevents binding of PI4P detector to PI4P. See Figure 3.

Figure 3. An example of a failed PI4P detection assay. Lipids were spotted onto the membrane together with the debris left over in the extraction step (the pellet at step C2). It prevented binding of PI(4)P Detector to PI4P and thus spots were detected as rings instead of circles.
Recipes
- Grinding buffer
0.33 M sorbitol
30 mM HEPES-KOH (pH 7.5)
2 mM EDTA
1x protease inhibitor cocktail
Autoclave without protease inhibitor cocktail
Protease inhibitor cocktail is added before use. - 80% or 40% Percoll
30 mM HEPES-KOH (pH 7.5)
0.33 M sorbitol
2 mM EDTA
Percoll (80% or 40%, v/v) - PBS
10 mM phosphate (pH 7.4)
150 mM NaCl
Sterilized by autoclave - PBST
PBS
0.1% (w/v) Tween-20
Acknowledgments
This protocol was adapted from the original work (Okazaki et al., 2015) to provide the detailed procedures. This work was supported by JSPS KAKENHI Grant Number 26840089.
References
- Klinkenberg, J. (2014). Extraction of chloroplast proteins from transiently transformed Nicotiana benthamiana leaves. Bio-protocol 4(18): e1238.
- Nakanishi, H., Suzuki, K., Kabeya, Y. and Miyagishima, S. Y. (2009). Plant-specific protein MCD1 determines the site of chloroplast division in concert with bacteria-derived MinD. Curr Biol 19(2): 151-156.
- Okazaki, K., Miyagishima, S. Y. and Wada, H. (2015). Phosphatidylinositol 4-phosphate negatively regulates chloroplast division in Arabidopsis. Plant Cell 27(3): 663-674.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Okazaki, K., Miyagishima, S. and Wada, H. (2016). Measurement of PI4P Levels in Intact Chloroplasts Isolated from Arabidopsis thaliana. Bio-protocol 6(3): e1719. DOI: 10.21769/BioProtoc.1719.
Category
Plant Science > Plant biochemistry > Lipid
Biochemistry > Lipid > Lipid measurement
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









