- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Preparation of Outer Membrane Vesicles from Myxococcus xanthus
Published: Vol 6, Iss 2, Jan 20, 2016 DOI: 10.21769/BioProtoc.1716 Views: 11447
Reviewed by: Arsalan DaudiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Extraction of Bacterial Membrane Vesicle and Phage Complex by Density Gradient Ultracentrifugation
Shangru Li [...] Tianyuan Jia
Aug 20, 2024 2054 Views

Mycobacterium smegmatis Ribosome Purification, Co-sedimentation, and Subunit Association Assay
Aneek Banerjee [...] Jayati Sengupta
May 20, 2025 1580 Views

Preparation and Negative Staining for Visualization of Cyanoglobule Lipid Droplets Using Transmission Electron Microscopy
Febri A. Susanto [...] Peter K. Lundquist
Dec 5, 2025 1311 Views
Abstract
Outer membrane vesicles (OMVs) represent a unique sub-cellular compartment of bacteria that may act as a scaffold for various extracellular activities, including intercellular signaling. Myxococcus xanthus (M. xanthus) is a predatory bacterium that engages in cell-cell behaviors such as fruiting body formation and contact dependent lysis of other microbes. The OMVs of M. xanthus have been shown to have an elaborate architecture of chains and tubes that can connect cells within a biofilm. These higher order OMV structures have been shown to contain proteins exchanged for community behaviors and small molecules that have antibiotic activities, and may help facilitate directed exchange. M. xanthus OMVs allow material transfer between neighboring cells for motility and predation.
Keywords: EPSMaterials and Reagents
- 50 ml plastic tubes for handling harvested cultures (VWR International, catalog number: 62406-200 )
- 0.45 μm syringe filters (VWR International, catalog number: 28145-477 )
- 0.22 μm syringe filters (VWR International, catalog number: 28144-050 )
- 30 ml syringes (VWR International, catalog number: 66064-760 )
- 1.5 ml Eppendorf tubes (VWR International, catalog number: 89213-152 )
- 200 mesh formvar coated TEM grids (Electron Microscopy Sciences, catalog number: EMS200-Cu )
- NuncTM MicroWellTM 96-Well Optical-Bottom Plates with Coverglass Base (Thermo Fisher Scientific)
- Myxococcus xanthus wild type strain DZ2 (UC Regents, Berkeley)
- Phosphate buffered saline (PBS) (VWR International, catalog number: 97064-158 )
- Fluorescent lipid dye FM 4-64 (Life Technologies, Molecular Probes®, catalog number: T-13320 )
Note: Currently, it is “Thermo Fisher Scientific, Molecular ProbesTM, catalog number: T-13320”. - Uranyl acetate (Electron Microscopy Sciences)
- 10 mM MOPS (pH 7.6)
- 2 mM MgSO4
- 10% (w/w) Bacto casitone
- 5% (w/w) Bacto yeast extract
- CYE Media (see Recipes)
Equipment
- Sterile, side arm 250 ml Erlenmeyer Flasks
- Shaking incubator (e.g., Thermo Fisher Scientific, model: MaxQ4000 )
- Vortexer
- Centrifuge with capacity for 25 ml cultures, 5,000 x g (Refrigeration not required)
- Ultra-centrifuge with capacity for 1 ml samples, 140, 000 x g required and refrigeration required. (e.g., GMI, Beckman Coulter, model: L8-70M )
- Ultra-centrifuge tubes (fit to machine specifications)
- -80 °C freezer
- Fluorescence plate reader (e.g., Tecan Trading AG, model: Vission-100 )
- Transmission Electron Microscope (Philips/FEI, model: 5350 NE Dawson Creek Drive ), capable of imaging negatively-stained samples at voltages ranging between 80 kv and 200 kV equipped with a 2 k x 2 k CCD camera
- Autoclave for sterilizing media and flasks (or access to sterile media and growth vessels).
Software
- Digital Micrograph software and 2 k x 2 k CCD camera (Gatan Inc.)
Procedure
- In order to obtain a cell free preparation of outer membrane vesicles and vesicle chains, M. xanthus strain DZ2 (wild type) cells were harvested from restreaks on CYE Petri plates, 3-21 days old and inoculated into 250 ml Erlenmeyer flasks with 25 ml of liquid CYE media (10:1 volume of flask:volume of culture ratio should be maintained for aerobic cultures of M. xanthus). A single 25 ml culture is sufficient for isolating ~200 μg of OMVs for further analysis.
- Cultures were grown with shaking at 200 rpm, 32 °C for 2 days to an OD600 = 2.0 (~2 x 109 cells/ml) in a Thermo Scientific MaxQ4000 shaking incubator.
- Cultures were transferred to 50 ml centrifuge tubes and vortexed for 30 sec to disperse aggregates and release vesicles. Note that for M. xanthus, large cell aggregates are common, and this duration of vortexing may not be needed when adapting to other bacterial species. Cultures should be handled at room temperature for steps 3-6, as cold temperature disrupts cell viability and impinges on fractionation.
- The vortexed culture was centrifuged for 10 min at 5,000 x g to pellet whole cells.
- The supernatant was filtered through a 0.45 μm syringe filter to remove any remaining cells.
- Filtrate was further centrifuged as in step 4, then passed through 0.22 μm syringe filters to remove debris.
- Cell free filtrate was then subjected to 140,000 x g centrifugation at 4 °C using an ultracentrifuge for 1 h to harvest outer membrane vesicles, vesicle chains and membrane tubes, that will be in the resulting pellet and resuspended in 1 ml of PBS.
- Samples were then saved at -80 °C or subjected immediately to analysis. Samples were analyzed by two methods described below to (a) monitor the purity of vesicle fractions by fluorescent lipid dye binding and (b) monitor the purity and quality of vesicle fractions by electron microscopy. Additional analyses can also be performed, for instance, proteomic analyses have indicated that the OMV protein fraction is consistent with little to no contamination from any other large protein complexes.
- Sample aliquots were mixed with fluorescent lipid dye FM 4-64 (purchased as a 100 μg dry stock, stored at -20 °C until use) to a final volume of 100 μl and final concentration of 16 μM. The relative concentration of vesicle samples was determined using black 96 well plates and a fluorescence plate reader by exciting at 515 nm, measuring emission at 640 nm. Five independent samples were analyzed for each biological preparation, as mixtures are heterogeneous. 16 μM FM 4-64 in PBS should be used as a negative control, while whole cell fractions can be utilized as positive controls. Fluorescence units may vary, but successful preparations should be 10-40 fold or higher than negative control readings. 1:1 Serial dilutions of samples may be required if signal saturation occurs. Yield increases with increasing cell density and with stable surface grown cultures (biofilms) relative to aerobic liquid cultures but ~200 μg or more is obtainable with this protocol.
- Negative-stained TEM samples were prepared by applying 3 μl of the OMV samples in 3 replicates to 200 mesh formvar coated TEM grids, allowing 3 min to settle and rinsing briefly in distilled water. A 2.0 % solution of Uranyl acetate was applied to cover the grid before immediately blotting off and air drying. Samples were imaged on a FEI CM200 microscope with a 2k x 2k Gatan CCD camera or on a JEOL 1200 microscope, operated at 200 kV or 120 kV, respectively, with quality judged by the presence of vesicles and vesicle chains and the absence of cells or cellular debris. For more detail on EM procedures see Remis et al. (2014a), for a video of EM procedures please see Rames et al. (2014b).
- Sample aliquots were mixed with fluorescent lipid dye FM 4-64 (purchased as a 100 μg dry stock, stored at -20 °C until use) to a final volume of 100 μl and final concentration of 16 μM. The relative concentration of vesicle samples was determined using black 96 well plates and a fluorescence plate reader by exciting at 515 nm, measuring emission at 640 nm. Five independent samples were analyzed for each biological preparation, as mixtures are heterogeneous. 16 μM FM 4-64 in PBS should be used as a negative control, while whole cell fractions can be utilized as positive controls. Fluorescence units may vary, but successful preparations should be 10-40 fold or higher than negative control readings. 1:1 Serial dilutions of samples may be required if signal saturation occurs. Yield increases with increasing cell density and with stable surface grown cultures (biofilms) relative to aerobic liquid cultures but ~200 μg or more is obtainable with this protocol.
Notes
Other EM analysis methods [SEM, resin-embedded sections TEM, cryo-EM of frozen-hydrated vesicle samples (Remis et al., 2014)] are also feasible for this assessment, but negative stain TEM is the simplest technique that provides this information. Light microscopy alone, due to its limited resolution, typically fails to provide accurate assessments of vesicle purifications.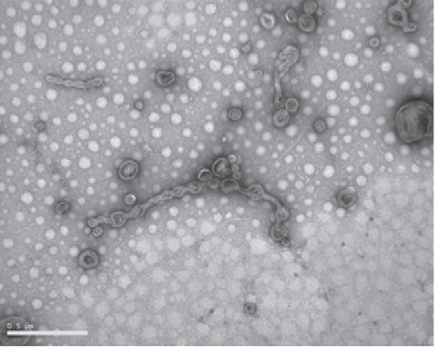
Figure 1. TEM image of M. xanthus OMVs (from Remis et al., 2014) showing isolated OMVs as well as OMV chains
Recipes
- CYE Media
10 mM MOPS (pH 7.6)
2 mM MgSO4
10% (w/w) Bacto casitone
5% (w/w) Bacto yeast extract
Sterilized by autoclaving
Acknowledgments
This protocol was adapted from previous work (Berleman et al., 2014; Palsdottir et al., 2009; Remis et al., 2014). This work was Supported by Lab directed research development funds from the Office of Biological and Environmental Research of the US Department of Energy under contract number DE-AC02-05CH11231 (to Manfred Auer) and the US Department of Energy VFP program (to James E. Berleman).
References
- Berleman, J. E., Allen, S., Danielewicz, M. A., Remis, J. P., Gorur, A., Cunha, J., Hadi, M. Z., Zusman, D. R., Northen, T. R., Witkowska, H. E. and Auer, M. (2014). The lethal cargo of Myxococcus xanthus outer membrane vesicles. Front Microbiol 5: 474.
- Palsdottir, H., Remis, J. P., Schaudinn, C., O'Toole, E., Lux, R., Shi, W., McDonald, K. L., Costerton, J. W. and Auer, M. (2009). Three-dimensional macromolecular organization of cryofixed Myxococcus xanthus biofilms as revealed by electron microscopic tomography. J Bacteriol 191(7): 2077-2082.
- Remis, J. P., Wei, D., Gorur, A., Zemla, M., Haraga, J., Allen, S., Witkowska, H. E., Costerton, J. W., Berleman, J. E. and Auer, M. (2014a). Bacterial social networks: structure and composition of Myxococcus xanthus outer membrane vesicle chains. Environ Microbiol 16(2): 598-610.
- Rames, M., Yu, Y. and Ren, G. (2014b). Optimized negative staining: a high-throughput protocol for examining small and asymmetric protein structure by electron microscopy. J Vis Exp(90): e51087.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Berleman, J. E., Zemla, M., Remis, J. P. and Auer, M. (2016). Preparation of Outer Membrane Vesicles from Myxococcus xanthus. Bio-protocol 6(2): e1716. DOI: 10.21769/BioProtoc.1716.
Category
Microbiology > Microbial cell biology > Organelle isolation
Cell Biology > Organelle isolation > Outer membrane vesicles
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










