- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Technique for the Measurement of in vitro Phospholipid Synthesis via Radioactive Labeling
Published: Vol 6, Iss 2, Jan 20, 2016 DOI: 10.21769/BioProtoc.1705 Views: 14636
Reviewed by: Neelanjan BoseDaniel KrausAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Liposome Flotation Assays for Phosphoinositide-protein Interaction
Helene Tronchere and Frederic Boal
Mar 5, 2017 19203 Views

Hyaluronan Isolation from Mouse Mammary Gland
Cornelia Tolg [...] Eva A. Turley
Jun 5, 2018 6089 Views

Enhanced Ribonucleoprotein Immunoprecipitation (RIP) Technique for the Identification of mRNA Species in Ribonucleoprotein Complexes
Saja A. Fakhraldeen [...] Caroline M. Alexander
Oct 5, 2022 2861 Views
Abstract
This is an assay designed to examine the radioactive phosphorous incorporation when the molecule is being synthesized, which means that only de novo synthesized phospholipids can be detected. Thus, with this technique it is possible to detect in vitro phospholipid synthesis under different required experimental conditions respect to controls (Guido and Caputto, 1990; Ferrero et al., 2014). There are different types of lipids. Among them we can find phospholipids, which contain glycerol esterified with two fatty acyl chains and a phosphate group that can also be bound to an organic molecule that acts as “hydrophilic head”, as shown in Figure 1 for the case of phosphatidylcholine. This structure confers amphipathic properties to lipid molecules that allow them to form lipid bilayers, making phospholipids the main components of biological membranes. 
Figure 1. Representation of phospholipid structure. Extracted from: http://bio1151.nicerweb.com/Locked/media/ch05/phospholipid.html
Materials and Reagents
- PYREX® 5 ml Rimless Kahn Culture Tubes (12 x 75 mm) (Corning, catalog number: 9820-12 )
- Scintillation vials (Sigma-Aldrich, catalog number: Z190527 )
- The enzymes and substrates are obtained as protein homogenates from homogenized cells or tissue (see Step 1 of the Procedure)
- Bio-Rad Protein Assay Dye Reagent Concentrate (Bio-Rad Laboratories, AbD Serotec®, catalog number: 5000006 )
- HEPES (Sigma-Aldrich, catalog number: H3375 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S7653 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9333 )
- D-(+)-Glucose (Sigma-Aldrich, catalog number: G8270 )
- Magnesium chloride (MgCl2) (Sigma-Aldrich, catalog number: M8266 )
- [γ32P]ATP (PerkinElmer, catalog number: BLU002001MC )
- Chloroform (Sigma-Aldrich, catalog number: C2432 )
- Methanol (Sigma-Aldrich, catalog number: 34860 )
- Trichloroacetic acid (TCA) (Sigma-Aldrich, catalog number: T6399 )
- Phosphotungstic acid (PTA) (Sigma-Aldrich, catalog number: P4006 )
- Non-aqueous liquid scintillation cocktail (PerkinElmer, catalog number: 1200-434 )
- Buffer HEPES (see Recipes)
- Reaction buffer (see Recipes)
- TCA-PTA 10-1 (% w/v) (see Recipes)
- TCA-PTA 5-0.5 (% w/v) (see Recipes)
- Carrier brain homogenate (see Recipes)
Equipment
- Tip sonicator (Branson Sonic Power Company, model: Sonifier B-12 ) or ULTRA-TURRAX®
- Spectrophotometer (Wavelength to be used: 595 nm) (Shimadzu Scientific Instruments, model: BioSpec-mini )
- Vortex (IKA® VORTEX 3) (Sigma-Aldrich, catalog number: Z654779 )
- Pipettes (PIPETMAN® L Starter Kit) (Gilson, catalog number: F167350 )
- Centrifuge (Cavourargetina, model: VT-3216Dx24 )
- Water thermostatic bath (Vicking, model: Masson D )
- Scintillation counter (WinSpectral, Wallac, PerkinElmer)
- Laboratory prepared with all the required equipment for safe receiving, storage, manipulation and waste processing of radioactive material, according to laws governing in each country
- Geiger counter (Ludlum Measurements, model: 3 survey meter )
Procedure
Note: To determine under which conditions lipid synthesis is linear with time, prepare different tubes and carry out the reaction at 37 °C for different times. If you are evaluating the capacity of modifying lipid synthesis of a given protein, you must also determine under which conditions lipid synthesis is linear with the concentration of your protein of interest.
The person that will carry on the experiments must have strict training with radioactive working, according to rules and governing laws.
- Incorporation of radioactive 32P into cell/tissue homogenates
- Grow the desired cell line with the required culture medium on a 10 cm dish until reaching 90% confluence.
Note: At this point, if you need your cells to be cultured under special conditions or subjected to a particular treatment, you should adequate the protocol to fit your needs. - Harvest cells with 300 µl of milliQ water or use a homogenizer or ULTRA-TURRAX® if the sample is obtained from a tissue, to obtain the protein homogenates according to experimental requirements.
- Sonicate on ice and measure total protein concentration using Bradford method or a similar one.
- Maintain samples on ice all the time.
- Prepare the reaction tubes. Each of them must contain a final volume of 80 µl, with 1x reaction buffer, 3 μCi of [γ32P] ATP and a protein concentration of 1.25 mg/ml. Consider that the samples must be measured at least in triplicate. For this, first aliquot in a Kahn tube the amount of protein homogenate necessary to reach the required protein concentration. Then, add the adequate volumes of the reaction buffer and the [γ32P] ATP to reach the concentrations mentioned above. As normally there are several conditions to be measured, and taking into account that they must be performed at least in triplicate, we find it convenient to prepare a master mix for all the tubes to avoid pipetting mistakes. For this, first aliquot the protein homogenate in the different tubes, then prepare a master mix with the adequate amounts of reaction buffer and [γ32P] ATP and aliquot it in the tubes. Finally, add enough milliQ water to reach the final volume.
Note: To simplify, it would help to normalize the protein concentration of all the samples so the volume of the master mix to be added is the same for all the tubes. Also, prepare a volume a bit larger of the master mix than the theoretical requirement, to compensate small volume losses or pipette miscalibrations. Adjust the final volume with milliQ water. - Vortex for 10 sec.
- Carry out the reaction at 37 °C for 60 min.
Note: At this time you can stop the reaction by freezing or you can continue with the purification steps. If you stop the reaction by freezing, the sample must be put in a bunker to avoid irradiation, according to working conditions with radioactive material. - Add 80 μl of TCA-PTA 10-1 (% w/v) to precipitate lipids and proteins. At this point, you should start seeing the imminent formation of a white precipitate. Vortex for 10 sec and centrifuge for 10 min at 3,500 rpm.
- Grow the desired cell line with the required culture medium on a 10 cm dish until reaching 90% confluence.
- Lipids precipitation and purification
- Discard the supernatant, which contains the free [γ32P] ATP that has not been incorporated, and vortex until the pellet is disaggregated.
- Add 1 ml of TCA-PTA 5-0.5 (% w/v) and vortex again for 10 sec. Centrifuge and repeat the pellet wash four times. Discard the supernatant after each round.
Note: If the amount of precipitate is too small, you can add 5 µl of brain homogenate, which will function as a carrier (see Figure 2).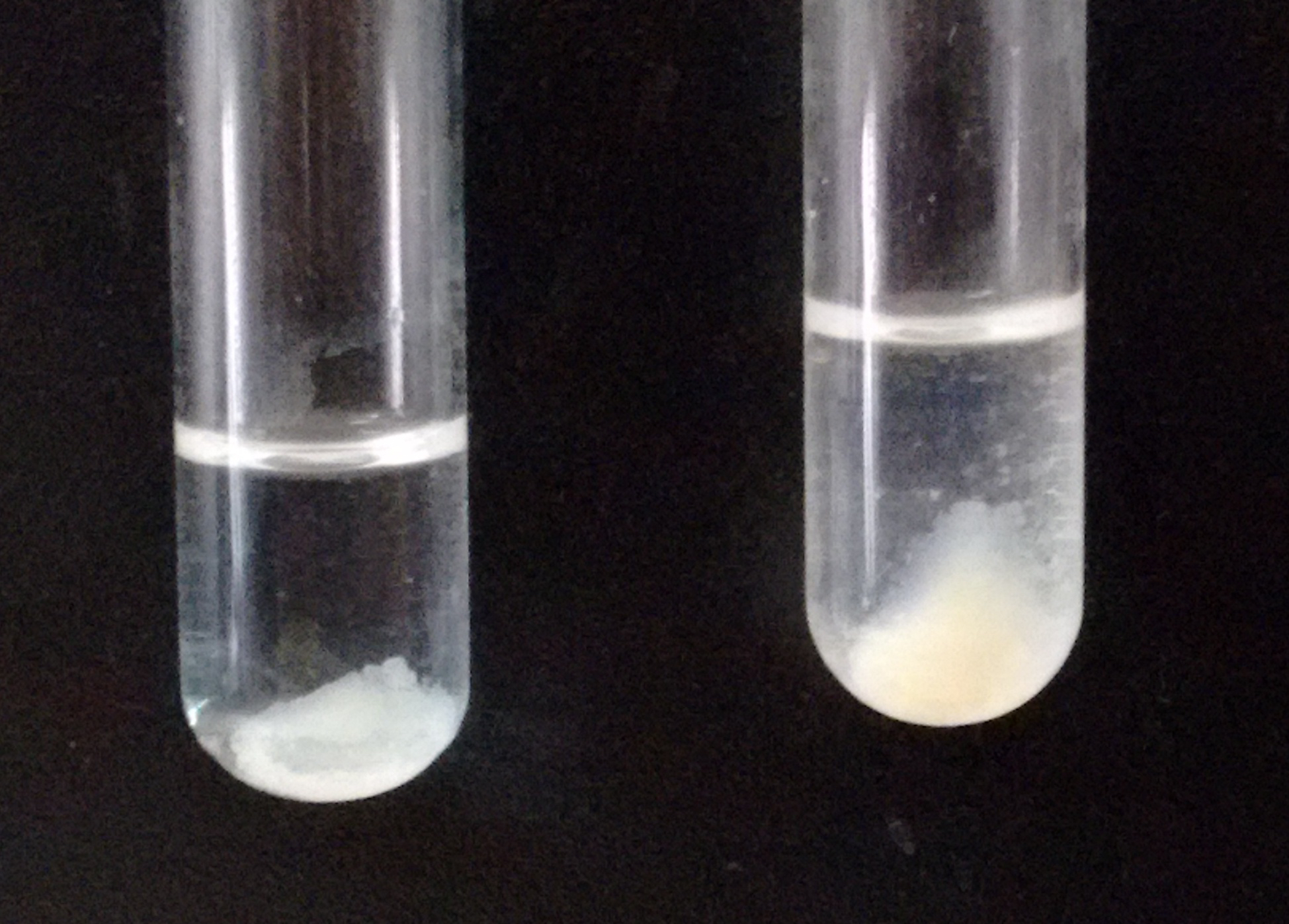
Figure 2. Example of the expected amount of precipitate (right tube) after adding the carrier brain homogenate. The tube on the left shows a precipitate considered too small. - Carry on a final wash with milliQ water, to favor lipid dissolution in the organic solvent (step B12).
- After centrifugation, discard the water supernatant of the last wash and vortex until the pellet is disaggregated.
- Add 1.5 ml of chloroform:methanol 2:1 and vortex for 10 sec. The chloroform dissolves the lipids and the methanol maintains the proteins precipitated.
- Centrifuge 10 min at 3,500 rpm to allow protein precipitation and separation from soluble lipids.
- Transfer 1.3 ml of the supernatant containing the marked lipids to a vial and dry in a water bath at 90 °C for 3 h or at room temperature overnight. When transferring the supernatant, be careful and do not pull precipitated proteins that may be marked with 32P.
Caution: As this step involves the transfer of a chloroform containing solution, glass pipettes previously equilibrated with chloroform should be used to avoid sample loss.
Note: There is a loss of 0.2 ml when transferring the supernatant to avoid disturbing the pellet.
- Discard the supernatant, which contains the free [γ32P] ATP that has not been incorporated, and vortex until the pellet is disaggregated.
- Measurement of 32P content in purified lipids
- Add 1 ml of non-aqueous scintillation cocktail, tap and vortex for 10 sec.
- Determine the levels of incorporated 32P in the lipids using a liquid scintillation counter. The reactions must be performed in triplicate, as minimum.
- Add 1 ml of non-aqueous scintillation cocktail, tap and vortex for 10 sec.
- Data analysis
- The results can be expressed as absolute values (as shown in Figure 3) or as relative values respect to a control condition taken as 100% of incorporation.
Representative data
Figure 3 is a representative example of data that indicates the type of results expected. In this case, the experimental condition measures the amount of phospholipid synthesis in the presence of c-Fos protein, its phosphorylated version or its mutants. In this regard, results are represented as amount of phospholipid labeling (Ferrero et al., 2012). 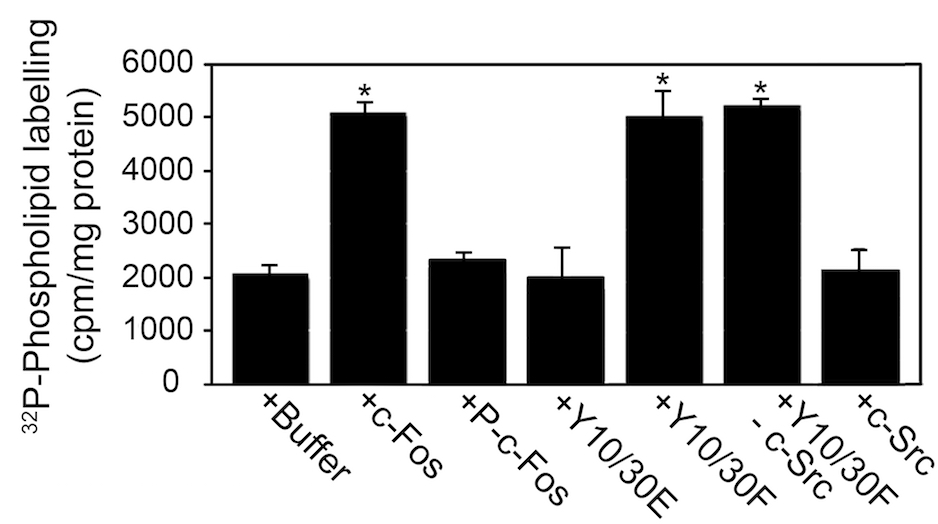
Figure 3. c-Fos phosphorylated by c-Src does not activate phospholipid synthesis. The capacity to activate phospholipid synthesis of recombinant c-Fos, c-Fos phosphorylated by purified c-Src (P-c-Fos), the phosphomimetic Y10/30E mutant of c-Fos and the non-phosphorylatable mutant of c-Fos Y10/30F was examined as described previously (Gil et al., 2004). Incubations were for 60 min at 37 °C. 32P-phospholipid quantification was performed as described previously (Guido and Caputto, 1990). Results expressed as c.p.m. of 32P incorporated into phospholipids/mg of protein are the mean±s.d. of three experiments performed in triplicate; *P<0.002 with respect to control (buffer) as determined by One Way ANOVA analysis. Y10/30F not incubated with c-Src and c-Src incubated without any added substrates were run as controls. Note that the presence of c-Src in the assays did not modify phospholipid synthesis.
Notes
If you decide to freeze after carrying out the reaction in step A6, don’t add the TCA-PTA 10-1 (% w/v) solution as the homogenate might agglomerate and precipitate together with non incorporated [γ32P] ATP molecules, which can lead to erroneous measurements.
Recipes
- Buffer HEPES (2 M, pH 7.5)
Dissolve 476.6 g HEPES in 800 ml of H2O
Adjust pH to 7.5 with the appropriate volume of concentrated NaOH
Add H2O to a final volume of 1 L - Reaction buffer (20x, 20 ml)Add milliQ water to a final volume of 20 ml
Reagent Quantity Concentration NaCl 3.2726 g 2,800 mM KCl 0.1491 g 100 mM MgCl2 0.019 g 10 mM Glucose 0.4036 g 112 mM HEPES buffer (2 M, pH 7.5) 12.8 ml
The buffer is stable for six months if stored at -20 °C. - TCA-PTA 10-1 (% w/v)
Weight 10 grams of TCA and 1 gram of PTA and add milliQ water to a final volume of 100 ml - TCA-PTA 5-0.5 (% w/v)
Aliquot 50 ml of the TCA-PTA 10-1 and add milliQ water to a final volume of 100 ml - Carrier brain homogenate
Euthanize an adult Wistar rat and excise the brain
Weight and prepare an homogenate of 1 g tissue per ml of milliQ water
Acknowledgments
We thank Dr. Beatriz L. Caputto for helpful discussions to adapt and modify the protocol form her previous work (Guido and Caputto, 1990). We also thank CONICET and FONCYT for funding.
References
- Guido, M. E. and Caputto, B. L. (1990). Labeling of retina and optic tectum phospholipids in chickens exposed to light or dark. J Neurochem 55(6): 1855-1860.
- Ferrero, G. O., Renner, M. L., Gil, G. A., Rodriguez-Berdini, L. and Caputto, B. L. (2014). c-Fos-activated synthesis of nuclear phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P(2)] promotes global transcriptional changes. Biochem J 461(3): 521-530.
- Ferrero, G. O., Velazquez, F. N. and Caputto, B. L. (2012). The kinase c-Src and the phosphatase TC45 coordinately regulate c-Fos tyrosine phosphorylation and c-Fos phospholipid synthesis activation capacity. Oncogene 31(28): 3381-3391.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Rodriguez-Berdini, L. and Ferrero, G. O. (2016). A Technique for the Measurement of in vitro Phospholipid Synthesis via Radioactive Labeling. Bio-protocol 6(2): e1705. DOI: 10.21769/BioProtoc.1705.
Category
Cancer Biology > General technique > Biochemical assays > Other compound
Biochemistry > Lipid > Lipid measurement
Cell Biology > Cell metabolism > Lipid
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











